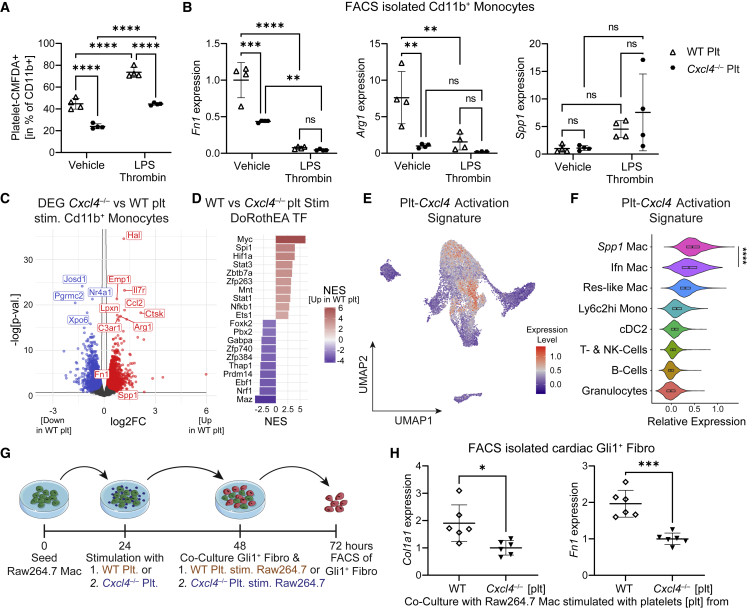
Figure 4.
Platelet-derived CXCL4 drives profibrotic Spp1+ macrophage activation
(A) Flow cytometric analysis of Platelet-CMFDA+CD11b+ platelet-monocyte aggregates after co-culture of WT PBMC with either CMFDA-positive WT or Cxcl4−/− platelets and stimulation with Vehicle or LPS and Thrombin (n = 4).
(B) RT-qPCR analysis for Arg1, Fn1, and Spp1 in sorted CD11b+ monocytes after WT or Cxcl4−/− platelet-induced activation of WT PBMC. Plt, platelets.
(C) Volcano plot showing differentially expressed genes in CD11b+ monocytes activated with either WT or Cxcl4−/− platelets (n = 4). p-val., p value; plt, platelets; stim., stimulated.
(D) DoRothEA transcription factor analysis of differentially expressed genes in CD11b+ monocytes co-cultured with either WT or Cxcl4−/− platelets.
(E) Expression of a platelet-Cxcl4 activation signature (top upregulated genes defined by an adjusted p value <0.01 and log2FC > 0.5 in WT versus Cxcl4−/− co-cultured CD11b+ monocytes) in cardiac immune cells plotted on the UMAP embedding shown in Figure 1A.
(F) Platelet-Cxcl4 activation signature in cardiac immune cells stratified by immune cell type.
(G) Experimental design of Gli1+-fibroblast co-culture with WT or Cxcl4−/− platelet-stimulated Raw264.7 macrophages. Mac, macrophages; Fibro, fibroblasts; stim, stimulated.
(H) RT-qPCR analysis of Col1a1 and Fn1 expression in Gli1+ cardiac fibroblasts after co-culture with WT or Cxcl4−/− platelet pretreated Raw264.7 macrophages as shown in (C) (n = 6).
For (A) and (B), a two-way ANOVA was computed using Tukey corrections. For (F) and (H), a two-tailed unpaired t test was performed. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001. See also Figure S5.