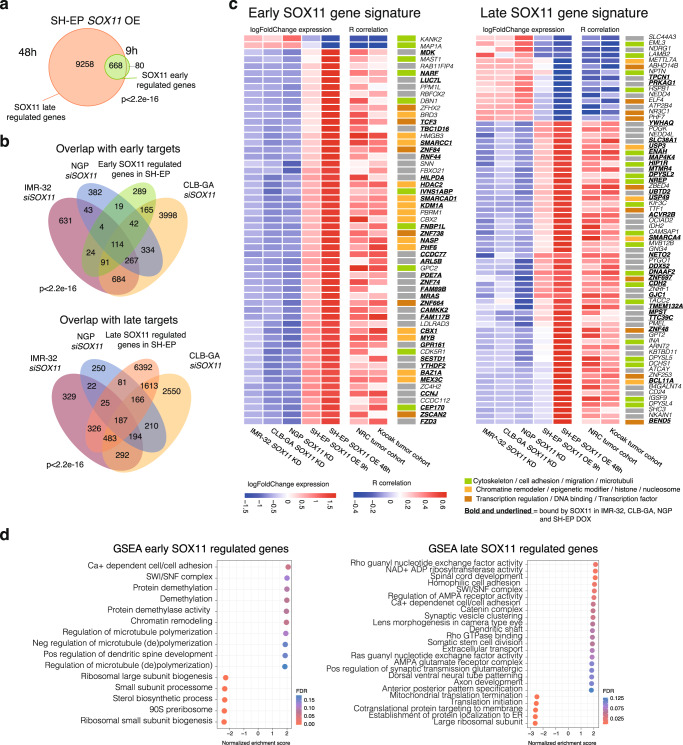
Fig. 4. The SOX11 regulated transcriptome is involved in epigenetic control, cytoskeleton and neurodevelopment.
a Overlap of genes perturbated in SH-EP upon SOX11 overexpression (OE) for 48 h and 9 h (adj p value <0.05) with differential genes upon 9 h overexpression being the SOX11 early regulated genes and differential genes upon 48 h but not 9 h overexpression being the SOX11 late regulated genes (one-tailed fisher test p-value <2.2e−16). b Overlap of genes perturbated in IMR-32, CLB-GA and NGP upon SOX11 knockdown for 48 h (adj p value <0.05) with respectively the SOX11 early and late regulated genes in SH-EP (one-tailed fisher test p-value <2.2e−16). c SOX11 early and late genes signature obtained by the overlap of differentially expressed genes upon SOX11 knockdown in IMR-32, CLB-GA and NGP with the early and late SOX11 regulated genes in SH-EP (adj p value <0.05, log FC > 0.5 or < −0.5) as well as with genes correlation with SOX11 expression in 2 different NB tumor cohorts (NRC GSE8504717, Kocak GSE4554718, p-value <0.05). A color next to each gene represents the involved pathways. Bold and underlined represents genes that are bound by SOX11 in IMR-32, CLB-GA, NGP and SH-EP after SOX11 overexpression for 48 h. d Top enriched genesets after doing GSEA analysis (http://www.gsea-msigdb.org/gsea/index.jsp, ontology gene sets C5) for SOX11 early and SOX11 late regulated genes. Depicted is the normalized enrichment score (NES, x-axis) and the false discovery rate (FDR, color). For Fig. 4c–d source data are provided as Source Data file.