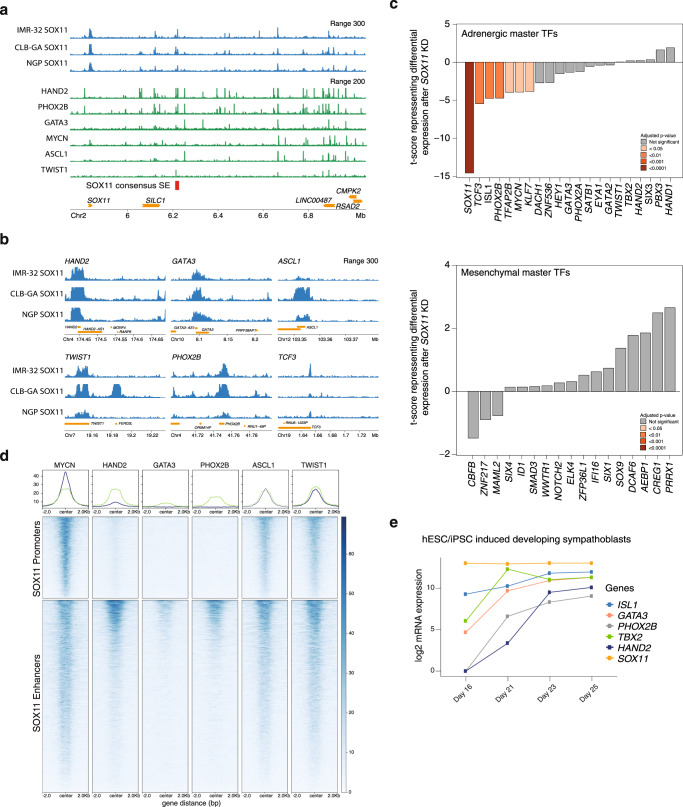
Fig. 6. SOX11 is a core regulatory circuitry transcription factor in adrenergic NB.
a Binding of SOX11 (CUT&RUN), HAND2, PHOX2B, GATA3 (GSE90683)4, MYCN, TWIST1 (GSE94822)46 and ASCL1 (GSE159613)47 and at the SOX11 locus and downstream enhancer landscape. Signal represents log likelihood ratio for the ChIP and CUT&RUN signal compared to input signal (RPM normalised). Super-enhancers of CLB-GA are annotated using ROSE (red bar) showing the SOX11 consensus super-enhancer (SE). b Binding of SOX11 to the PHOX2B, HAND2, GATA3, ASCL1, TCF3 and TWIST1 locus. Signal represents log likelihood ratio for the ChIP and CUT&RUN signal compared to input signal (RPM normalised). c Tscore representing differential expression of MES and ADRN core regulatory circuitry transcription factors (TFs) in RNA-sequencing data after SOX11 knockdown (KD) in IMR-32, CLB-GA and NGP. Significant adjusted p-values are indicated with coloured bars. Statistical testing was done using the empirical Bayes quasi-likelihood F-test. d Heatmap profiles −2 kb and +2 kb around the summit of SOX11 CUT&RUN peaks in IMR-32, grouped for promoters or enhancers (homer annotation). On these regions HAND2, PHOX2B, GATA3 (GSE90683)4, MYCN, TWIST1 (GSE94822)46 and ASCL1 (GSE159613)47 ChIP data is mapped and ranked according to the sums of the peak scores across all datasets in the heatmap. e ISL1, GATA3, PHOX2B, TBX2, HAND2 and SOX11 (log2) expression during induced differentiation of hPSC cells along the sympatho-adrenal lineage. Expression levels depicted starting from day 16, upon sorting the cells for SOX10 expression indicating cells committed to truncal neural crest cells, and followed-up during sympatho-adrenal development until day 25. For Fig. 6c and e source data are provided as Source Data file.