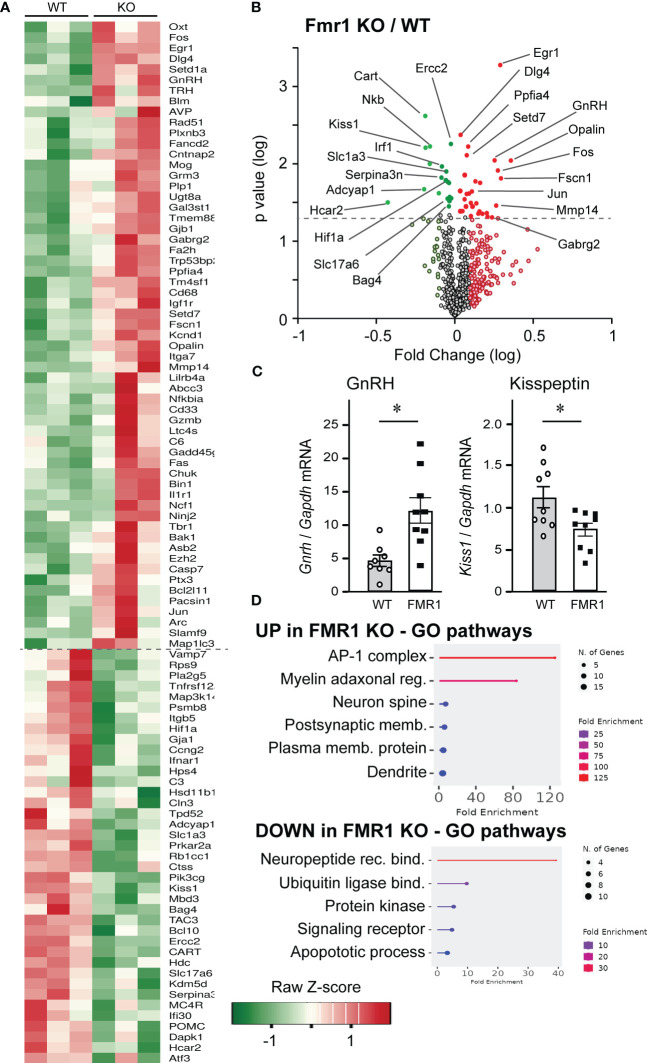
Figure 7.
Nanostring analysis demonstrates significant changes in hypothalamic gene expression in Fmr1 KO mice. (A) Hypothalami from 3 mice per group were dissected and 50 ng RNA used in Nanostring analysis. Heatmap indicates gene expression changes in Fmr1 KO hypothalami that are <0.8-fold and >1.2-fold over WT. (B) Data were plotted as log fold change on x-axis vs. log p value on y-axis, and dashed line indicates significance. Red indicates genes that are increased in KO compared to WT mice, while green indicates genes that are decreased in KO compared to WT. Genes below the dotted line, light red and light green did not reach significance. (C) qPCR of the hypothalamus confirms changes in GnRH and kisspeptin expression. Each point represents one animal and bars represent group average. Statistical significance is indicated with *, determined by t-test followed by Tukey’s post hoc test. (D) Gene ontology pathway analysis indicates upregulated (top) and downregulated (bottom) pathways in the Fmr1 KO mice.