Significance
B cells are an important antigen presenting cell subset in the thymus that require activation to mediate T cell central tolerance. The process of thymic B cell activation, termed licensing, results in isotype class-switch recombination and upregulation of self-antigens and antigen presentation pathways in B cells. In this work, we show thymic B cells express the type III interferon(IFN) receptor and rely on type III IFN for licensing. Absence of the type III IFN receptor on B cells results in impaired T cell central tolerance, as illustrated by a reduction in regulatory T cell generation and altered Treg cell T cell receptor repertoire, demonstrating the critical role for licensed B cells in regulatory T cell development.
Keywords: thymic B cells, central tolerance, Treg cell selection, type III IFN
Abstract
The activation of thymic B cells is critical for their licensing as antigen presenting cells and resulting ability to mediate T cell central tolerance. The processes leading to licensing are still not fully understood. By comparing thymic B cells to activated Peyer’s patch B cells at steady state, we found that thymic B cell activation starts during the neonatal period and is characterized by TCR/CD40-dependent activation, followed by immunoglobulin class switch recombination (CSR) without forming germinal centers. Transcriptional analysis also demonstrated a strong interferon signature, which was not apparent in the periphery. Thymic B cell activation and CSR were primarily dependent on type III IFN signaling, and loss of type III IFN receptor in thymic B cells resulted in reduced thymocyte regulatory T cell (Treg) development. Finally, from TCR deep sequencing, we estimate that licensed B cells induce development of a substantial fraction of the Treg cell repertoire. Together, these findings reveal the importance of steady-state type III IFN in generating licensed thymic B cells that induce T cell tolerance to activated B cells.
Thymic central tolerance is the process where developing T cells interact with self-antigens in the thymus to generate a diverse MHC-restricted T lymphocyte repertoire that protects against pathogens while limiting responses to self-antigens (1). T cell central tolerance is mediated by T cell receptor (TCR) recognition of self-antigen–MHC complexes presented by antigen presenting cells (APCs) in the thymus (2, 3). Thymic APCs express unique types and quantities of self-antigens, are in distinct anatomical locations, and demonstrate different costimulatory protein expression (2, 3). Thus, the various populations of thymic APC likely play unique roles in mediating T cell central tolerance.
One such unique APC subset is thymic B cells. Thymic B cells represent only a small proportion of cells in the thymus but are present early in life and increase with age (4). These B cells undergo a unique activation process, termed thymic B cell licensing (5). Licensing is most likely initiated by CD4SP recognition of self-peptide:MHCII presented by B cells, as it is absent in TCRa-deficient mice, mice that have a fixed nonself-reactive TCR, and mice that lack MHCII on B cells (5, 6). Self-reactive CD4SP thymocytes express CD40L and the delivery of CD40L-CD40 signaling is also essential for thymic B cell licensing (5). This T:B interaction results in isotype class-switched B cells with enhanced antigen presentation capabilities and the expression of unique self-antigens partially driven by AIRE (5). B cells presenting self-antigens in the thymus can drive clonal deletion (5, 6). Less is understood about B cell-mediated thymic Treg cell selection, but both BAFF-Tg and WT mice showed reduced Treg cell selection in the absence of B cells (7, 8). It is unknown whether B cell-mediated Treg cell selection requires B cell licensing. Nonetheless, these works demonstrate thymic B cells induce T cell central tolerance in thymocytes specific for self-antigens expressed by activated B cells (9).
How thymic B cells become licensed is still incompletely understood. Expression of a polyclonal TCR repertoire is necessary for thymic B licensing, but it is not clear whether differentiated T helper cells are required for the process (6). The thymus has a small population of differentiated IL-21 producing thymocytes that could be influencing B cell activation (10), as IL-21 is known to promote B cell proliferation and germinal center (GC) formation (11), but it is unknown whether these cells contribute to licensing. Traditionally, differentiated follicular helper T cells (Tfh) and germinal centers (GCs) are associated with B cell class-switch recombination (CSR) and somatic hypermutation (SHM) in peripheral lymphoid organs (12). However, recent evidence shows CSR occurs independently of GC and/or Tfh (13, 14). Investigation of the intrinsic and extrinsic requirements for thymic B cell licensing will help to further understand their contribution to thymic central tolerance.
Here, we investigated the development of licensed thymic B cells and their role in thymic central tolerance. Licensed class-switched thymic B cells were distinct from B cells in Peyer’s patches (PPs) as they did not show a traditional GC immunophenotype or transcriptome. Instead, licensed thymic B cells rapidly developed during the first weeks of life and were found to be independent of GC and Tfh. When transcriptionally compared with other activated peripheral B cells, licensed thymic B cells expressed high amounts of interferon-stimulated genes (ISGs). Surprisingly, thymic B cell activation and resulting licensing was dependent on type III interferon (IFN) signaling in B cells. Loss of type III IFN signaling resulted in alterations in B cell activation, CSR, ISG expression, and reduced Treg cell development. To confirm the impact of licensed B cells on T cell central tolerance, TCR repertoire sequencing was performed, showing a large portion of Treg cells require class-switched B cells for development. Thus, class-switched, licensed thymic B cell are essential for thymic Treg cell selection.
Results
Class-Switched Thymic B Cells Exhibit a Unique Phenotype.
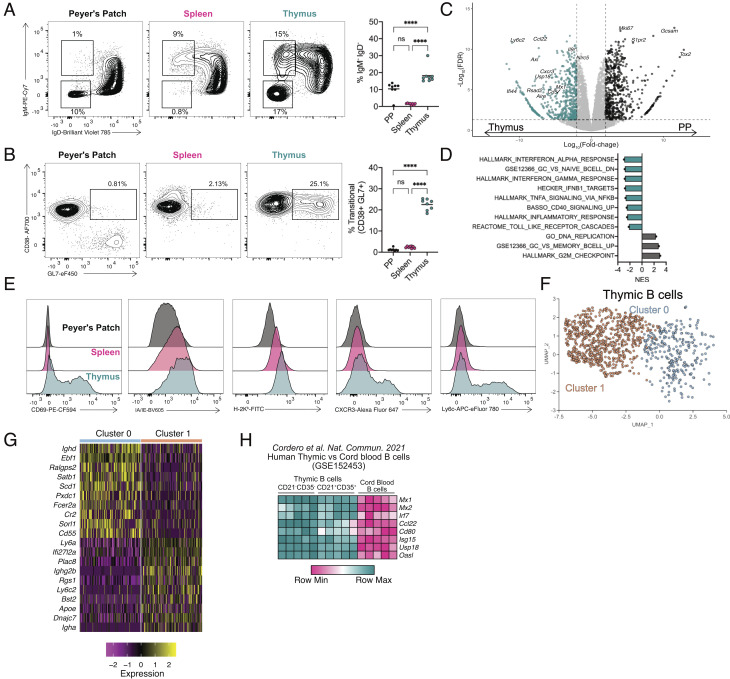
To better define the unique aspects of thymic B cell activation, we first set out to compare thymic B cells with splenic and Peyer’s patch (PP) B cells. Both PP and thymic B cells have high frequencies of class-switched IgM−IgD− B cells at steady state, while B cells from the spleen show little CSR (Fig. 1A). As reported previously (6), class-switched thymic B cells were predominantly switched to IgG2b, IgA, and IgG2c isotypes (SI Appendix, Fig. S1). Thymic and splenic B cells also showed a population of IgM+IgDlo B cells that were absent in the PP (Fig. 1A). Further immunophenotyping was performed with CD38 and GL7 to identify naïve/memory (CD38+GL7−), activated memory precursor (CD38+GL7+), and germinal center (GC) (CD38−GL7+) B cells. Like B cells in PPs, thymic B cells up-regulated the germinal center activation marker GL7. However, they did not down-regulate CD38 (Fig. 1B), as is typical for GC B cells (15). Such CD38+GL7+-activated memory B cell precursors were previously shown to be generated independent of Bcl6 but could give rise to GC or memory B cells (15). Activated memory B cell precursors were further shown to be formed by CD40 signaling, but independently of full T cell help, which may explain their presence in the thymus where CD4SP thymocytes can provide CD40 signaling but are unlikely to differentiate fully and provide help to generate a GC response, as self-reactive CD4SP thymocytes undergo apoptosis. Taken together, there is a large population of class-switched B cells present in the thymus, but they do not express typical immunophenotypic markers associated with a GC response.
Fig. 1.
Class-switched thymic B cells exhibit a unique phenotype. (A) Flow cytometry of B cells from Peyer’s patch (Left), spleen (Middle), and thymus (Right) showing IgM and IgD expression, with number adjacent to outlined areas identifying proportion of isotype class-switched (IgM−IgD−) and activated IgM+IgDlo B cells. Graph on right shows quantification of isotype class-switched B cells (IgM−IgD−). (B) Flow cytometry of total B cells from the Peyer’s patch (Left), spleen (Middle), and thymus (Right) showing CD38 and GL7 expression with number adjacent to outlined areas identifying proportion of activated memory B cell precursors. Graph on Right shows quantification of activated memory B cell precursors (CD38+GL7+). (C) Volcano plot representation of RNA-Seq data comparing isotype class-switched thymic and PP B cells showing Log2 fold-change by -Log10 FDR. Significantly up-regulated genes (FDR <= 0.05) are identified in the thymus (blue, FC < −2) and PP (black, FC > 2). (D) Gene set enrichment analysis normalized enhancement score (NES) for ranked RNA-Seq data. Negative NES are pathways overrepresented in thymic B cells; positive NES pathways overrepresented in PP B cells. (E) Flow cytometry histograms for CD69, I-A/I-E, H2-Kb, CXCR3, and Ly6C expression of B cells in the PP (black, Top), spleen (pink, Middle), and thymus (blue, Bottom). (F) Uniform Manifold Approximation and Projection (UMAP) plot for thymic B cells. (G) Heatmap representation of top 10 differentially expressed genes in subsampled thymic B cells from cluster 0 and 1. (H) Heatmap representation of log2-transformed counts per-million (CPM) of individual ISG mRNA in CD21−CD35− and CD21+CD35+ thymic B cells compared with cord blood B cells. Each symbol (A and B) represents an individual mouse; small horizontal lines indicate the group mean. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, and ****P ≤ 0.0001 (A and B One-way ANOVA with Tukey’s multiple comparisons test).
To compare the transcriptomes of thymic and PP B cells, IgM−IgD− B cells from the thymus and PP were sorted and RNA-seq was performed on the isolated RNA, identifying 512 genes up-regulated in thymic B cells, and 546 up-regulated in PP B cells (Fig. 1C). Gene set enrichment analysis (GSEA) showed thymic B cells were significantly enriched for pathways associated with inflammation such as IFN, TNF-a, and TLR signaling, while PP B cells showed enrichment of proliferation and GC response pathways (Fig. 1D). Flow cytometry confirmed higher expression of CD69, MHCI, MHCII, Ly6C. and CXCR3 (Fig. 1E), and qPCR confirmed higher expression of Aire, Ccl22, Cd40, Cd80, Ly6c2, Mx1, and Irf7 in thymic B cells (SI Appendix, Fig. S1B). To define the heterogeneity of the thymic B cell population, thymic B cells were analyzed by single-cell RNA-Seq (scRNA-Seq), revealing two distinct clusters (Fig. 1F). Differential gene expression analysis suggested these two clusters could be characterized as unswitched naïve/memory B cells (Ighd in cluster 0) or activated and isotype class-switched B cells (Ighg2b, Igha in cluster 1) (Fig. 1G and SI Appendix, Fig. S1C). Cluster 1 was also defined by several interferon-stimulated genes (ISGs) (Ly6a, Ifi27l2a, Ly6c2) (Fig. 1G and SI Appendix, Fig. S1C). It is important to note that cluster 1 was not comprised solely of isotype class-switched B cells, as some cells still expressed Ighm even though they lacked Ighd (SI Appendix, Fig. S1C). This suggests that IgD downregulation represents an activated state of B cells even though they were not isotype switched. Last, we investigated human thymic B cells to determine if they shared a similar activation as mouse. Comparison of previously published human neonatal thymic and peripheral (cord blood) B cells revealed human thymic B cells were strongly enriched for many of the same interferon pathways identified in mouse thymic B cells (SI Appendix, Fig. S1D) (16). Analysis of individual differentially regulated genes in mouse thymic B cells showed a similar expression pattern as human thymic B cells (Fig. 1H). Thus, thymic B cells from mice and humans demonstrate a phenotype associated with inflammatory signaling in the absence of a traditional GC transcriptome.
CD40L Drives a Burst of B Cell Licensing in Early Life.
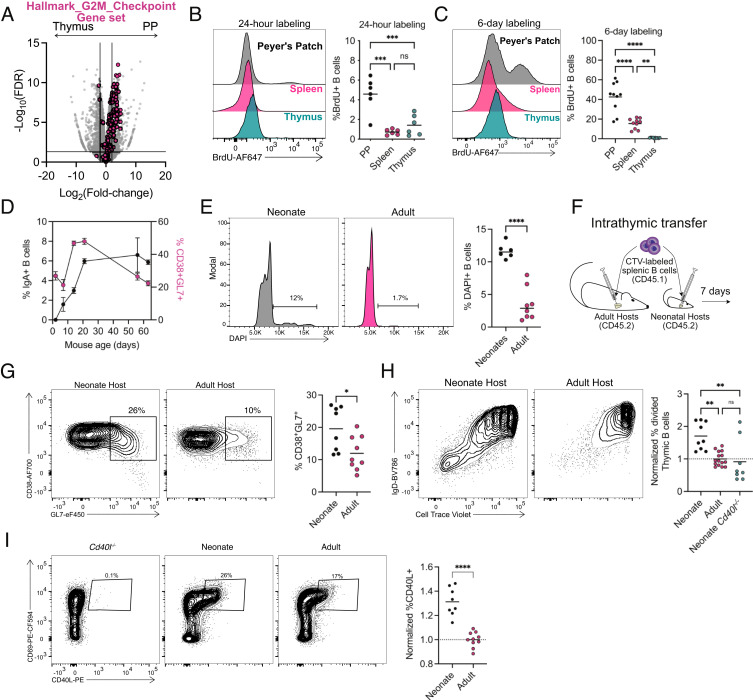
Thymic B cells are highly differentiated with a large population of class-switched B cells at steady state. The differentiation of peripheral B cells during immune responses is tightly coordinated with proliferation (17, 18); however, thymic B cells showed a distinct absence of genes associated with proliferation when compared with PP B cells (Fig. 2A). We confirmed the low proliferative state of thymic B cells by BrdU labeling in adult mice for either 24-h (Fig. 2B) or 6-d (Fig. 2C). At both timepoints, thymic B cells incorporated significantly less BrdU than PP B cells (Fig. 2 B and C). Previous work reported thymic B cells undergo activation and CSR within days of birth in both mice and humans (19). Thus potentially, the proliferative differences between thymic and PP B cells could be due to differences in the timing of B cell activation and CSR. Analysis of thymic B cells from birth revealed the presence of CD38+GL7+ B cells already at 2 d of age and class-switched B cells shortly thereafter, which peaked in frequency at 21 d of age (Fig. 2D). The frequency of isotype-switched B cells remained stable into adulthood, while the frequency of CD38+GL7+ B cell was reduced after 3 wk (Fig. 2D). This suggested that thymic B cells may undergo a burst of activation early in life. To test if proliferation of thymic B cells occurred in this period, cell cycle analysis using DAPI was performed, demonstrating a greater frequency of neonatal thymic B cells in S/G2M when compared with the adult (Fig. 2E). This confirms that a larger fraction of thymic B cells are undergoing activation, differentiation, and proliferation during neonatal life.
Fig. 2.
Increased CD40L expression drives a burst of B cell licensing in early life. (A) Volcano plot representation of RNA-Seq data comparing isotype class-switched thymic and PP B cells (gray) with overlaid “Hallmark_G2M_Checkpoint” gene set (pink). (B) Flow cytometry histogram measuring BrdU incorporation after 24-h labeling (2-mg intraperitoneal injection) of B cells in the PP (black, Top), spleen (pink, Middle), and thymus (blue, Bottom). Graph of right shows quantification of BrdU-positive B cell after 24 h of labeling. (C) Flow cytometry histogram measuring BrdU incorporation after 6 d of labeling (0.8 mg/mL in drinking water) of B cells in the PP (black, Top), spleen (pink, Middle) and thymus (blue, Bottom). Graph on right shows quantification of BrdU-positive B cell after 6 d of labeling. (D) Time course of thymic B cell IgA isotype CSR (black, left axis) and CD38+GL7+ (pink, right axis) activation. Points represent mean (±SEM). (E) Flow cytometry histogram measuring DAPI in thymic B cells from neonatal (Left) and adult (Right). Graph on right shows quantification of DAPI-positive B cells. (F) Diagram of intrathymic transfer experiments into adult or neonatal hosts. (G) Flow cytometry of splenic B cells following intrathymic transfer showing CD38 by GL7 expression in neonatal (5 to 7 d old) (Left) and adult (6 to 8 wk old) (Right) host mice. Graph on right shows quantification of transferred CD38+GL7+ B cells. (H) Flow cytometry of splenic B cells following intrathymic transfer showing IgD and Cell Trace Violet (CTV) expression in neonatal (5 to 7 d old) (Left) and adult (6 to 8 wk old) (Right) host mice. Frequency of divided intrathymic transferred donor B cells normalized to the frequency of divided B cells in adult hosts. (I) Flow cytometry of CD4SP thymocytes (CD4+ CD8α- CD25-GITR-CD44-) showing CD40L by CD69 with number adjacent to outlined areas identifying proportion of CD40L+. The graph on right shows the normalized frequency of CD40L+ CD4SP thymocytes to adult CD40L frequency on the right. Each symbol (B, C, E, and G–I) represents an individual mouse; small horizontal lines indicate the group mean. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001 and ****P ≤ 0.0001 (B, C, H One-way ANOVA with Tukey’s multiple comparisons test, E, G, I Two-tailed unpaired Student’s t test).
To further test whether the neonatal thymic environment enhances the licensing of thymic B cells, we used an adoptive transfer model (Fig. 2F). Splenic B cells were labeled with cell proliferation dye and intrathymically injected into host mice. Transfer of splenic B cells resulted in upregulation of CD69, MHCII, GL7, and Ly6C in the thymus, as well as isotype switching, similar to that seen in thymic B cells at steady state (Fig. 2G and SI Appendix, Fig. S2A). Transfer of splenic B cells into day 5 to 7 neonatal thymi showed increased proliferation when compared with transfer into adult thymi (Fig. 2H). Noticeably, the most divided cells in both hosts were primarily composed of IgD− B cells (Fig. 2H). Analysis of the IgD− CTV− transferred B cells revealed these cells were composed of both IgM+ (activated) and IgM− (isotype switched) B cells demonstrating activation, licensing, and CSR occurred in cells undergoing multiple rounds of division (SI Appendix, Fig. S2 B and C), and a higher fraction of B cells underwent this process in neonatal recipients than in adult.
The neonatal thymic environment is distinctive as it generates unique TCRs that have higher self-reactivity due to lack of N-nucleotide additions (20, 21). Thymocyte CD40L expression is dependent on self-reactivity in CD4SP thymocytes (22); therefore we hypothesized that increased neonatal thymocyte self-reactivity leads to increased CD40L expression and B cell licensing. Indeed, CD40L expression was higher on CD4SP thymocytes from neonates (Fig. 2I). and B cell proliferation after transfer into neonatal mice was reduced in neonatal Cd40l−/− hosts (SI Appendix, Fig. S2B). Together, these results suggest that as mice age and CD40L expression on CD4SP is reduced, less thymic B cells are newly licensed, which results in a reduced frequency of actively proliferating B cells. As the numbers of thymic B cells do not decrease as mice age, this model suggests the majority of activated thymic B cells maintain residency in the thymus. Indeed, parabiosis studies suggested thymic B cells are primarily resident in the thymus (9, 23), supporting the hypothesis that thymic B cells are activated early in life and become resident.
Finally, during neonatal life, B cells are primarily of fetal origin (24, 25). If most of thymic B cell activation occurs in the neonatal period and those B cells are retained in the thymus, then the population of thymic B cells in adult mice would include more B cells of fetal origin. To address this, we analyzed previously reported BCR repertoire data from adult thymic versus splenic B cells for N-nucleotide addition frequency. We observed that the frequency of BCRs with N-nucleotide additions was lower in thymic B cells (both switched and unswitched) than in splenic B cells, albeit not absent (SI Appendix, Fig. S3A). This supports the idea that B cell licensing is more active in the neonatal period.
Thymic B Cell Activation and Class-Switching Is Not Dependent on Tfh Cells.
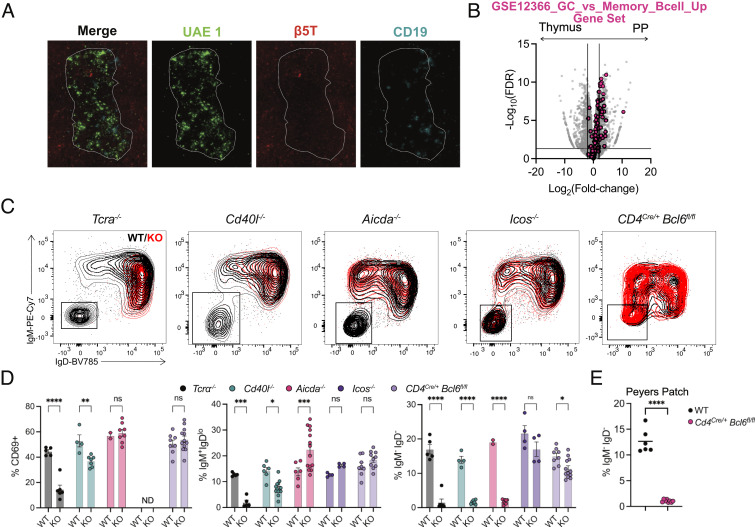
Although the contribution of T cells to thymic B cell licensing is well established (5), it is unclear whether Tfh cells or GC structures are required for thymic B cell CSR. Tfh cells are a T cell subset specialized for providing B cell help during peripheral immune responses and are necessary for the generation of GC B cells (26). However, not all B cells need to enter a germinal center or interact with Tfh cells to undergo CSR (13, 14). We first investigated where thymic B cells were localized using immunofluorescence. We found only small B cell clusters, generally located at the corticomedullary junction (Fig. 3A). There was no evidence of an organized GC structure, and B cell clusters were rare outside of the thymus medulla. Lack of a transcriptional GC signature in thymic B cells was further confirmed by GSEA, showing isotype class-switched thymic B cells were transcriptionally more similar to memory B cells and showed reduced expression of GC defining genes such as Bcl6, Pcna7, Pms2, Brca1, Fas, and S1pr2, with increased expression of Bcl2 when compared with class-switched PP B cells (Fig. 3B) (27). To investigate the contribution of Tfh cells in B cell licensing, we examined Icos−/− and CD4Cre/+Bcl6fl/fl mice, which are unable to generate Tfh cells (28). Thymic B cells from mice lacking αβ T cells or CD40L have reduced activation (CD69 upregulation and IgD downregulation) and CSR as previously shown (6) (Fig. 3 C and D). Thymic B cells from mice lacking AID show initial activation but no CSR, as expected. However, thymic B cells from both Icos−/− and CD4Cre/+Bcl6fl/fl mice showed normal levels of activation and CSR, despite profoundly reduced GC B cells in the Peyer’s patch in CD4Cre/+Bcl6fl/fl mice (Fig. 3E). In summary, thymic B cell licensing is a germinal center-independent activation process.
Fig. 3.
Thymic B cell class switching does not require TFH. (A) Immunofluorescence of the thymus showing UAE-1 (green), β5T (red), and CD19 (cyan). Medullary regions are outlined by a white dotted line. (B) Volcano plot representation of RNA-Seq data comparing isotype class-switched thymic and PP B cells (gray) with overlaid “GSE12366_GC_VS_MEMORY_BCELL_UP” gene set (pink). (C) Flow cytometry overlays of thymic B cells showing IgM by IgD from WT (black) and respective knockout mice (TCRα−/−, Aicda−/−, Cd40l−/−, Icos−/−, and CD4Cre/+ Bcl6fl/fl). (D) Quantification of activated B cells (CD69+), isotype class-switched (IgM−IgD−) and activated IgM+IgDlo thymic B cells. (E) Quantification of isotype class-switched (IgM−IgD−) B cells in Peyer’s patch of WT and Cd4Cre/+ Bcl6fl/fl mice. Each symbol (D and E) represents an individual mouse; small horizontal lines indicate the group mean ± SEM. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001 and ****P ≤ 0.0001 [(D) Two-way ANOVA with Sidak’s multiple comparisons test, (E) Two-tailed unpaired Student’s t test].
Type III IFN Drives Thymic B Cell Licensing.
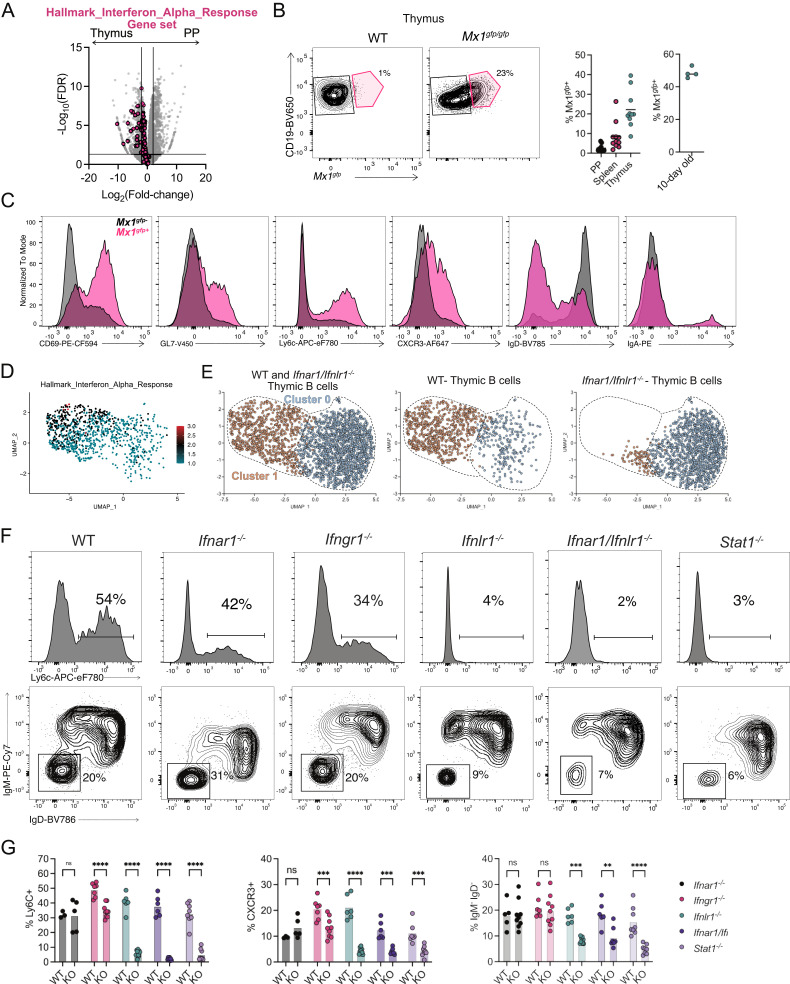
IFN signaling pathways were highly enriched in thymic B cells when compared with B cells from the PP (Fig. 1D and 4A). IFNs can enhance B cell survival, activation, and production of antibodies during autoimmunity and in response to viral challenges (29). Type I and III IFNs are expressed by epithelial cells in the thymus at steady state (30, 31). Thus, IFN could potentially impact thymic B cell function. To investigate the role of IFN in thymic B cell licensing, we analyzed B cells from Mx1gfp/gfp mice, where GFP is expressed in cells actively responding to IFN (32). In 6- to 8-wk-old mice, 20% of thymic B cells expressed Mx1 in Mx1gfp/gfp mice while B cell expression of GFP in peripheral tissues was low (Fig. 4B). Thymic B cells from 10-d-old mice showed ~50% Mx1gfp expression, supporting our finding of increased thymic B cell activation in early life (Fig. 4B). GFP+ thymic B cells showed greater activation (CD69, GL7), upregulation of Ly6C and CXCR3, and CSR (IgD, IgA) (Fig. 4C). Thus, IFN signaling in thymic B cells is associated with activation and CSR. To identify global changes occurring in B cells due to IFN signaling, the previous scRNA-Seq data were further analyzed. A module score for the “Hallmark IFN Alpha Response” gene set was generated and applied to the thymic B cells, finding the activated B cells in cluster 1 were strongly enriched for IFN signaling (Fig. 4D). Within this scRNA-Seq dataset, thymic B cells from WT and Ifnar1−/−/Ifnlr1−/− mice were mixed and labeled with hashtag-oligos (HTOs) to identify cellular origin. Thymic B cells from Ifnar1−/−/Ifnlr1−/− mice were used to ablate signaling from both type I and III IFNs. Thymic B cells from Ifnar1−/−/Ifnlr1−/− mice were poorly represented within cluster 1 that was composed of activated and isotype switched B cells (Fig. 4E). Therefore, IFN signaling is necessary to drive the differentiation of activated and class-switched B cells in the thymus. Next, to determine the contribution of individual IFN types in the processes of activation and CSR, the thymic B cell phenotype was analyzed in mice deficient in type I IFN signaling (Ifnar1−/−), type II IFN signaling (Ifngr1−/−), type III IFN signaling (Ifnlr1−/−), both type I and III IFN signaling (Ifnar1−/−/Ifnlr1−/−) or mice deficient for Stat1 (Stat1−/−), a transcription factor necessary for the transmission of type I, II and III IFN signaling (33–35). Analysis of thymic B cells showed that expression of the ISGs Ly6C and CXCR3 were dependent on type III IFN but not type I IFN receptors and showed partial dependence on type II IFN (Fig. 4 F and G). CSR was reduced by deficiency of Ifnlr1, but not Ifnar1 or Ifngr1. It is unclear what role type II IFN is playing in thymic B cells’ ISG regulation, but it does not impact the frequency of thymic B cell licensing (Fig. 4 F and G). In summary, type III IFN drives activation, expression of ISGs, and isotype class switching in thymic B cells.
Fig. 4.
Type III IFN drives thymic B cell licensing. (A) Volcano plot representation of RNA-Seq data comparing isotype class-switched thymic and PP B cells (gray) with overlaid “Hallmark_Interferon_Alpha_Response” gene set (pink). (B) Flow cytometry of thymic B cells showing CD19 and Mx1gfpin WT and Mx1gfp/gfp mice with number adjacent to outlined areas identifying proportion of Mx1gfp+ thymic B cells. Graphs on right showing quantification of Mx1gfp+ PP, splenic and thymic B cells in adult mice, and thymic B cells from 10-d-old mice (C) Flow cytometry histogram overlays of Mx1gfp+ (pink) and Mx1gfp− (black) thymic B cells showing expression of CD69, GL7, Ly6C, CXCR3, IgD, and IgA. (D) UMAP plot of thymic B cells with module score identifying cells expressing genes identified in the “Hallmark_Interferon_Alpha_Response” gene set. (E) UMAP plots of thymic B cells derived from WT and Infar1−/−/Inflr1−/− hosts identified by HTO. (F) Flow cytometry histograms showing Ly6C expression (Top) or isotype class-switched (IgM−IgD−) thymic B cells derived from WT, Ifnar1−/−, Ifngr1−/−, Ifnlr1−/−, Ifnar1−/−/Ifnlr1−/−, and Stat1−/− mice. Numbers adjacent to outlined areas identifying the proportion of cells within the gate. (G) Quantification of Ly6C+, CXCR3+, and isotype class-switched (IgM−IgD−) thymic B cells from WT, Ifnar1−/−, Ifngr1−/−, Ifnlr1−/−, Ifnar1−/−/Ifnlr1−/,− and Stat1−/− mice. Each symbol (B and G) represents an individual mouse; small horizontal lines indicate the group mean. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001 and ****P ≤ 0.0001 [(G) Two-way ANOVA with Sidak’s multiple comparisons test].
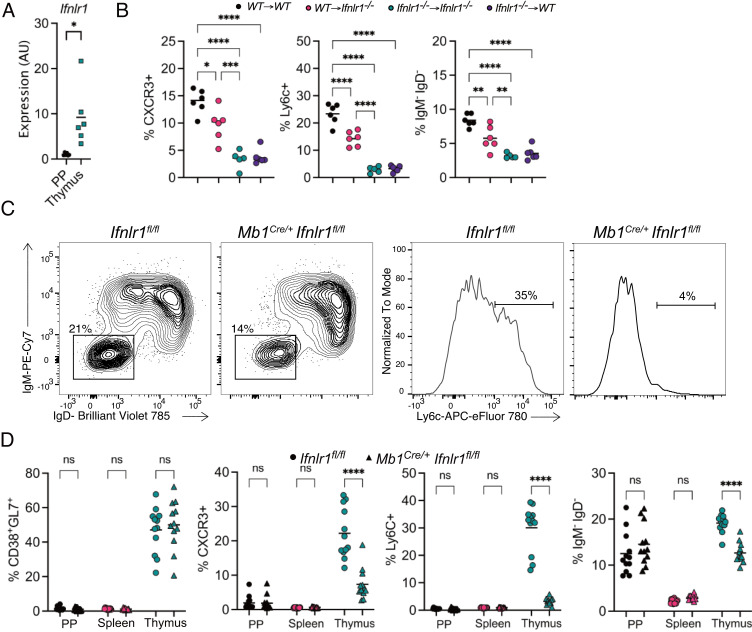
Requirement for Type III IFN Receptor is B Cell Intrinsic.
We sought to determine whether thymic B cells respond directly to type III IFN. Ifnlr1 expression was measured by RT-qPCR in B cells from the thymus and PP, finding greater expression of the receptor in thymic B cells (Fig. 5A). Next, we generated bone marrow chimeric mice where either the hematopoietic donor cells, recipients or both lacked Ifnlr1 expression. Thymic B cell expression of ISGs CXCR3 and Ly6C, as well as isotype class-switching, were dependent on hematopoietic expression of Ifnlr1 (Fig. 5B). Interestingly, when Ifnlr1 was absent only on recipient cells, B cells showed an intermediate level of Ly6C and CXCR3 expression and class switching (red symbols in Fig. 5B), suggesting a partial effect in radioresistant cells. Next, thymic B cells were analyzed in CD4Cre/+Ifnlr1fl/fl and Mb1Cre/+Ifnlr1fl/fl mice to determine if B cell licensing was intrinsically controlled by type III IFN on T or B cells. Thymic B cells from CD4Cre/+Ifnlr1fl/fl mice showed no changes in B cell licensing (SI Appendix, Fig. S4A) while thymic B cells from Mb1Cre/+Ifnlr1fl/fl showed significant reduction in CXCR3 and Ly6C expression, as well as isotype class switching (Fig. 5 C and D). The absence of Ifnlr1 in B cells impacted thymic B cell CSR while demonstrating no impact on CSR in Peyer’s patch (Fig. 5D), nor did PP B cells express CXCR3 or Ly6C. Isotype class-switched thymic B cells were further characterized for individual isotypes, finding reduction of all isotypes, suggesting type III IFN impacts thymic CSR in general and not switching to a single isotype (SI Appendix, Fig. S4B). Thus, type III IFN directly drives thymic B cells’ ISG upregulation and CSR.
Fig. 5.
Requirement for type III IFN receptor is B cell intrinsic. (A) RT-qPCR of Ifnlr1 in sorted IgM−IgD− B cells from the PP (black) and thymus (blue) normalized to Gapdh. (B) Proportion of thymic B cells from mixed bone marrow chimeras with the following phenotypes: Ly6C+, CXCR3+, and IgM−IgD−. (C) Flow cytometry of thymic B cells from Ifnlr1fl/fl and Mb1Cre/+Ifnrl1fl/fl showing isotype class-switched (IgM−IgD−) (Left) and Ly6C expression (Right). Numbers adjacent to outlined areas identifying the proportion of cells within the gate. (D) Quantification of the proportion of CD38+GL7+, CXCR3+, Ly6C+, and isotype class-switched (IgM−IgD−) B cells from Ifnlr1fl/fl and Mb1Cre/+Ifnrl1fl/fl mice located in the PP, spleen, or thymus. Each symbol (A, B, and D) represents an individual mouse; small horizontal lines indicate the group mean. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001 and ****P ≤ 0.0001 [(A) Two-tailed unpaired Student’s t test, (B) One-way ANOVA with Tukey’s multiple comparisons test, (D) Two-way ANOVA with Sidak’s multiple comparisons test].
Type III IFN Drives Tolerogenic Activity of Thymic B Cells.
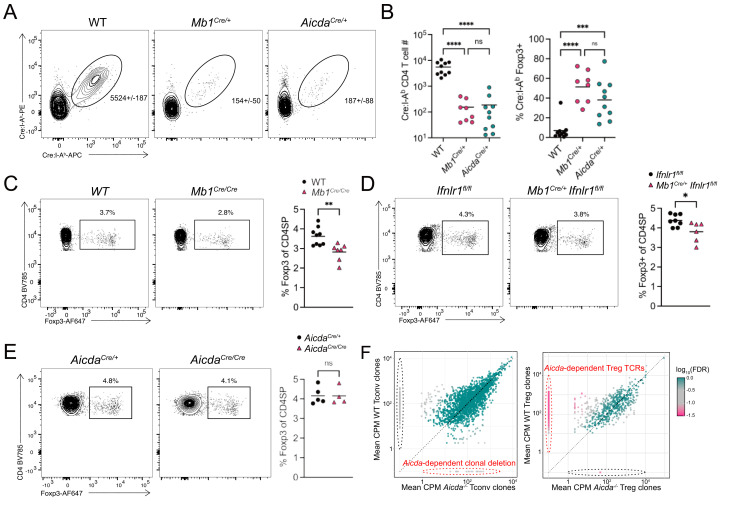
Thymic B cells can mediate CD4SP thymocyte clonal deletion (5, 9, 36, 37) and Treg cell generation (7, 8). To verify that B cell self-antigens induced T cell central tolerance, we examined tolerance to a model self-antigen—Cre—when its expression was restricted to B cells. Previous work showed that Cre:I-Ab-specific CD4 T cell immune responses in Cre-expressing mice could be used to study both deletion and Treg cell generation to tissue-restricted antigens (38). WT, Mb1Cre/+, and AicdaCre/+ mice were immunized with the immunodominant Cre61–71 peptide emulsified in CFA (Fig. 6A). Cre protein was expressed as a self-antigen in all B cells (Mb1Cre/+) or only in isotype class-switched B cells (AicdaCre/+). Analysis of the immune response following immunization showed reductions in total Cre:I-Ab-specific CD4 T cell numbers and an increased frequency of Cre:I-Ab-specific Foxp3+ Treg cells (Fig. 6 A and B). The total expansion of Cre:I-Ab-specific CD4 T cells and the proportion of Foxp3+ Treg cells was similar between Mb1Cre/+ and AicdaCre/+ mice, demonstrating licensed B cells can induce both clonal deletion and Treg cell induction.
Fig. 6.
Type III IFN signaling in thymic B cells is required for Foxp3+ regulatory T cell selection. (A) Flow cytometry of live CD19− CD11b− CD11c− F4/80− TCRβ+ CD4+ T cells showing Cre:I-Ab-PE by Cre:I-Ab-APC to identify Cre61–71:I-Ab-specific CD4 T cells. Numbers adjacent to outlined areas identify the average number of cells in the gate ± the SD of all samples. (B) Quantification of the absolute number (Left) and Foxp3+ regulatory T cell frequency (Right) of Cre61–71:I-Ab-specific CD4 T cells 7 d following Cre61–71/CFA subcutaneous immunization. (C) Flow cytometry of CD4SP thymocytes showing Foxp3 by CD4 in WT (C57BL/6) and Mb1Cre/Cremice (Left). Numbers adjacent to outlined areas identify the proportion of cells within the gate. Quantification of the proportion of Foxp3+ regulatory T cells in the CD4SP thymocyte population (Right). (D) Flow cytometry of CD4SP thymocytes showing Foxp3 by CD4 in Ifnlr1fl/fl and Mb1Cre/+Ifnrl1fl/fl mice (Left). Numbers adjacent to outlined areas identify the proportion of cells within the gate. Quantification of the proportion of Foxp3+ regulatory T cells in the CD4SP thymocyte population (Right). (E) Flow cytometry of CD4SP thymocytes showing Foxp3 by CD4 in AicdaCre/− and AicdaCre/Cremice (Left). Numbers adjacent to outlined areas identify the proportion of cells within the gate. Quantification of the proportion of Foxp3+ regulatory T cells in the CD4SP thymocyte population (Right). (F) Comparison of the mean CDR3 clonotype counts per million reads mapped in Tconv (Left) and Treg cells (Right) from WT or AicdaCre/Cre Tcra+/− TclibTg Foxp3eGFP mice (n = 4 mice per genotype). The Log10(FDR) of for each CDR3 peptide counts per million is shown by heatmap, and CDR3 clonotypes with an FDR ≤ 0.05 are represented by square points and CDR3 clonotypes with an FDR >0.05 are represented by circular points. CDR3 clonotypes identified in red dotted ellipses represent clonal deletion (Left) and regulatory T cell generation (Right) dependent upon class-switched B cells. Each symbol (B–E) represents an individual mouse; small horizontal lines indicate the group mean. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001 and ****P ≤ 0.0001 [(C–E) Two-tailed unpaired Student’s t test, (A) Two-way ANOVA with Sidak’s multiple comparisons test].
To further study the role of licensed thymic B cells on thymocyte selection, we first confirmed previous reports that B cell-deficient (Mb1Cre/Cre) mice had reduced proportions of polyclonal thymic Treg cells (Fig. 6C). The previous study showing that mice lacking B cells had reduced proportions of Treg cells did not distinguish nascent thymic Treg cells from Treg cells that recirculated back to the thymus (7, 8). Thus, we distinguished developing Treg cells from recirculating thymic Treg cells using the marker CD73 (39). Nascent Treg cells (CD73−) were decreased in B cell-deficient mice as well, demonstrating that developing Treg cells require B cells (SI Appendix, Fig. S5A). As type III IFN drives thymic B cell activation and licensing, we hypothesized type III IFN would also impact thymocyte Treg cell selection. Analysis of developing thymocytes in Mb1Cre/+ Ifnlr1fl/fl mice found decreased Treg cell development when only B cells lacked type III IFN signaling (Fig. 6D). As in total B cell-deficient mice, both total and nascent Foxp3+ Treg cells were reduced when type III IFN signaling was removed from B cells (Fig. 6D and SI Appendix, Fig. S5B). Thus, thymic B cell activation requires type III IFN signaling to optimally induce thymic Treg cell development.
To determine if the lack of licensed, class-switched B cells was driving the Treg cell selection changes in Mb1Cre/+ Ifnlr1fl/fl mice, we next analyzed Treg cell development in mice lacking class-switched B cells (AicdaCre/Cre). Interestingly, these mice did not show reduced Foxp3+ Treg cell frequency when compared with control mice (AicdaCre/+) (Fig. 6E). As mice lacking isotype CSR B cells (AicdaCre/Cre) did not show altered thymic Treg cell frequencies but were able to induce Treg cell selection in the Cre-model antigen system, we next tested whether isotype CSR thymic B cells altered the Treg cell TCR repertoire. We crossed AicdaCre/Cre mice to mice expressing a fixed TCRβ transgene- TclibTg- to reduce the repertoire diversity to ensure sufficient sequencing coverage (40, 41). We also crossed these mice to the Tcra−/− mouse to generate Tcra+/− experimental mice so that each T cell could only express and be selected by a single αβ TCR. Conventional CD4SP (Tconv) and Treg thymocytes were flow sorted from the thymus of individual mice and the Tcra locus sequenced using arm-PCR technology. Unique CDR3 sequences were parsed, and the data were filtered to include only those sequences present at >10 cpm and in at least three of four individual mice analyzed. Comparison of TCR clonotypes in WT and AicdaCre/Cre mice for Tconv (Left) and Treg (Right) cells found a subset of clones were significantly overrepresented in WT Treg cells compared with the AicdaCre/Cre Treg cells (Fig. 6F), suggesting class-switched B cells are required for the selection of these clones. Tconv cell clone selection was not statistically different between WT and AicdaCre/Cre mice (Fig. 6F), although given the larger, more diverse repertoire of Tconv cells, analysis of more animals would be required to conclude that the repertoires are not different. Altogether, these data demonstrate licensed B cells play a critical role in Treg cell repertoire selection.
Discussion
Here, we have identified a requirement for type III IFN in generating tolerogenic thymic B cells. Surprisingly, type III IFNs are constitutively expressed in the thymus (31). While type III IFN was shown to impact thymic epithelial cell antigen presentation and CD8 T cell clonal deletion through increasing MHC Class I levels, our data define type III IFNs impact on Foxp3+ Treg cell selection mediated by thymic B cells. We found that thymic B cell licensing starts soon after birth, driven in part by the increased CD40L expression by neonatal thymocytes, which are more self-reactive than adult thymocytes (20, 21). Along with CD40 signaling, type III IFN receptor signaling is required on thymic B cells to induce full activation, isotype class-switch recombination, and licensing. Reduction in thymic B cell activation in the absence of type III IFN signaling resulted in a poorly tolerogenic APC population with reduced ability to generate Foxp3+ Treg cells. The nascent Treg cell population was reduced by approximately 16% in B cell-deficient mice, suggesting that B cells are a major thymic APC involved in generating Treg cells. Surprisingly, the Treg cell reduction was almost as large (11%) in Mb1Cre/+Ifnlr1fl/fl mice, suggesting that type III IFN-induced licensing of B cells is critical to their ability to generate Treg cells, possibly through the upregulation of MHCII and unique self-antigen expression. While there were no changes in the global frequency of thymic Treg cells in mice lacking B cell isotype CSR (AicdaCre/Cre), analysis of the TCR repertoire found it to be altered by class-switched B cells. Altogether these findings suggest that thymic B cell activation plays a major role in the selection and shaping the Treg cell TCR repertoire.
Type III IFNs signaling through the IFNLR1/IL-10Rβ heterodimeric receptor plays an important role in antiviral defenses of barrier tissues (42). Our data provide definitive evidence that mouse B cells can express IFNLR1 and respond to type III IFN. Responsiveness to type III IFN is thought to be controlled by expression of IFNRL1, as IL-10Rβ is widely expressed across multiple cell types while IFNLR1 expression is more restricted (42, 43). Previous work found that peripheral B cells did not directly respond to type III IFN but instead showed that type III IFN acted as an extrinsic regulator of B cell activation through recruitment of APCs (44). However, others have shown type III IFN can intrinsically act on peripheral B cells, leading to the suppression of plasmablast formation in response to malaria infection in mice, or increasing plasmablast differentiation in human cells (45, 46). Thus, it seems that under some physiological conditions, B cells can express IFNLR1 and type III IFN signaling directly impacts B cells. In our work, only thymic B cells were affected by the loss of the type III IFN receptor, while peripheral B cell activation and isotype CSR in Peyer’s patches were unaffected. This may be due to poor expression of IFNLR1 in peripheral B cells, as demonstrated by isotype class-switched B cells in the PPs. The unique CD4SP thymocyte-thymic B cell interaction or the thymic microenvironment may drive expression of IFNLR1, allowing thymic B cells to respond to thymic epithelial cell produced type III IFN.
Medullary thymic epithelial cells (mTEC) express type III IFNs, while both medullary and cortical thymic epithelial cells (cTECs), thymic B cells, and thymic cDC1 express IFNLR1 and are responsive to type III IFN signaling (31). Like type I IFN, type III IFN production by mTECs is partially dependent on AIRE expression (31). Analysis of transcriptome data from mTEC fate tracking experiments shows the highest type III IFN production in AIRE+ mTECHi (MHCII+) cells, suggesting differentiated mTECs are necessary for type III IFN production (47). However, AIRE control of type III IFN production may be direct, like a tissue-restricted antigen, indirect through the upregulation of a gene product or transcript that induces type III IFN or due to the requirement of AIRE in mTEC differentiation (48). Further studies of how type III IFN production is regulated by AIRE will be important to understand the control of type III IFN, thymic B cell licensing, and resulting T cell tolerance.
The initial step in B cell licensing is driven by a CD40L+ CD4SP thymocyte recognition of self-antigen:MHCII displayed by thymic B cells (5). The delivery of the CD40L-CD40 signaling initiates activation of thymic B cells, resulting in isotype CSR, but requires the support of type III IFN to undergo CSR in many thymic B cells. Once licensed, thymic B cells present AIRE-dependent antigens (5), as well as epitopes likely derived from Ig and ISGs (49), and drive Foxp3+ Treg cell selection. It is interesting to note that in this model of thymic B cell licensing, two thymocytes with distinct self-reactive TCRs are required. The first self-reactive thymocytes delivering CD40L recognizes antigens presented by resting B cells to initiate licensing, while the second self-reactive thymocyte recognizes antigens presented by the licensed thymic B cell, resulting in Treg cell selection. The fate of the first self-reactive thymocyte delivering CD40L signaling is unknown. However, if self-reactive thymocytes can interact with resting thymic B cells, then why is licensing essential for T cell central tolerance generation? We hypothesize there are two roles for licensing. The first is that licensing induces upregulation of MHCII and costimulatory molecules, increasing capabilities of B cells to act as APC. The second is that licensing likely induces broad changes in self-antigen expression and processing in thymic B cells. It is known that B cell activation alters antigen processing to increase the efficiency of presenting Ig-captured antigens, thus altering the B cell self-peptidome (50, 51). Thus, licensing increases the efficacy of B cell-induced selection, while also producing new self-antigens derived from activated B cell self-antigens.
We confirmed thymic B cell licensing occurs within the first days of life and is greatly enhanced in the neonatal thymus due to increased CD4SP CD40L expression. While we found the neonatal environment enhances thymic B cell licensing, there may be other intrinsic and extrinsic factors that regulate licensing. Thymic B cells in the neonatal period may be uniquely suited to become licensed. During neonatal life, B cells in peripheral organs are primarily of fetal origin and bone marrow-derived B cells do not displace them until the end of the neonatal period (24, 25). Parabiosis and our BrdU studies support the hypothesis of early thymic B cell seeding by B cells, as there is proliferation and differentiation of B cells early in life, but by adulthood thymic B cells are resident and largely quiescent. Thus thymic B cells may be skewed toward those of fetal origin, and the observation of reduced N-nucleotide additions is consistent with this. However, peripheral B cells can enter the thymus in adulthood, as intravenously transferred B cells are found licensed in the thymus, even though this process does not seem to be efficient (5). Furthermore, fetal B cell progenitors commonly produce B-1 cells, but B-1 cells are phenotypically distinct from thymic B cells (CD5+, diffuse CD43+ and CD11b−) (9). Finally, fetal liver and bone marrow showed comparable reconstitution of the thymic B cell niche in lethally irradiated or Rag-1−/− host adult recipients (5, 9). Thus, fetal B cell progenitors do not seem to be uniquely susceptible to licensing, but rather there is a supportive neonatal environment, created at least in part by increased CD4SP CD40L. IFN expression in the thymus may also contribute to the supportive neonatal environment. IFN expression by mTEC is partially regulated by AIRE (30, 31), and mTEC cellularity peaks following the neonatal period (52, 53), potentially increasing the amount of IFN present within the thymus. In this scenario, the neonatal thymus has increased ability to induce B cell licensing through higher concentrations of IFN and increased expression of CD40L on CD4SP. Experiments to determine the influence of IFN expression in neonatal and adult mice will be informative to further address these hypotheses.
B cell activation in peripheral lymphoid organs results in isotype CSR and SHM, with the later occurring within germinal centers (12, 14). This B cell activation and germinal center formation is primarily generated in response to foreign antigens and is tightly regulated by Tfh and T follicular regulatory (Tfr) cells to make high-affinity antibodies and avoid overt autoreactivity (12, 26, 54, 55). Yet in the thymus, B cell licensing occurs in response to self-antigens in the presence of “sterile” inflammation driven by interferons. Even though there are differences between B cell activation in the thymus and periphery, it is likely thymic B cells will present and generate tolerance to the same B cell-derived self-antigens that are generated and displayed during a B cell immune response to a foreign antigen following infection. Thus, the outcome of licensed thymic B cell-mediated thymic central tolerance may serve to minimize autoreactivity during humoral immune responses, either in preventing T cell-mediated destruction of activated B cells, or through the regulation of activated B cells during CSR and SHM by the generation of Tfr cells.
The self-antigens generated by type III IFN-driven thymic B cell licensing are likely exclusively presented by this APC population, as loss of Ifnlr1 on thymic B cells results in a large reduction in Treg cell selection. Though we hypothesize the AIRE-expressing thymic B cell would be reduced when Inflr1 is absent, we do not believe the loss of tissue-restricted antigens regulated by AIRE are solely driving changes in T cell central tolerance. Instead, we propose that two other groups of self-antigens may be presented by type III IFN-licensed thymic B cells that drive T cell selection: ISGs and Ig. Based on scRNA-seq, the thymic B cells that require type III IFN for licensing express high amounts of ISGs, and likely present epitopes derived from these self-antigens to developing thymocytes. For example, thymic B cells in mice lacking Ifnlr1 show loss of Ly6c and CXCR3 expression (Fig. 4F) and would not present epitopes derived from these proteins. At this time, it is not known which ISGs are solely produced in type III IFN-licensed B cells, but future studies will be helpful to define different IFNs role in controlling ISG expression and potentially identify unique selecting ISG-derived epitopes. The other source of self-antigen driven by B cell licensing are likely those derived from class-switched Ig. TCR repertoire analysis demonstrated mice lacking Aicda have a large Treg cell repertoire defect, with many of these epitopes likely being derived from Ig. Self-antigens derived from Ig can be presented by thymic B cells (49) and we hypothesize they are driving Treg cell development. Future studies will focus on identifying T cell receptors that recognize Ig- and ISG-derived epitopes and determine if they contribute to the development of autoimmunity reported in aged mice lacking Ifnlr1 (31).
Materials and Methods
Mice.
5- to 8-wk-old male and female, age-matched mice were used for all experiments. C57BL/6NCrl and B6.SJL-PtprcaPepcb/BoyCrl were purchased from Charles River Laboratories. B6.129S2-Ifnar1tm1Agt/Mmjax (Strain #:028288, Ifnar1−/−), B6.129S(Cg)-Stat1tm1Dlv/J (Strain #:012606, Stat1−/−), B6.129S7-Ifngr1tm1Agt/J (Strain #:003288, Ifngr1−/−), B6.C(Cg)-Cd79atm1(cre)Reth/EhobJ (Strain #:020505, Mb1Cre), B6.Cg-Tg(Cd4-cre)1Cwi/BfluJ (Strain #:022071, Cd4Cre), B6.Cg-Foxp3tm2Tch/J (Strain #:006772, Foxp3eGFP), and B6.129P2-Aicdatm1(cre)Mnz/J (Strain #: 007770, AicdaCre/Cre) mice were purchased from The Jackson Laboratory. B6.Cg-Mx1tm1.1Agsa/J (Mx1gfp) were kindly provided by A. García-Sastre (Icahn School of Medicine at Mount Sinai) and have been described previously (32). Ifnlr1tm1.1Svko (Ifnrl1fl/fl) and Ifnlr1tm1.2Svko (Ifnrl1−/−) mice were kindly provided by Sergei V Kotenko (Rutgers New Jersey Medical School) and have been described previously (35). Tg(Tcrb51–11.5)AR1251Ayr (TClibTg) mice have been described previously (41). Mice were maintained and bred under specific pathogen-free conditions at the University of Minnesota. All animal experiments were approved by the Institutional Animal Care and Use Committee of the University of Minnesota. All animals were maintained under specific pathogen-free conditions at the University of Minnesota.
Immunizations.
Mice were administered a subcutaneous injection of 50 μL 1:1 CFA(Sigma-Aldrich):PBS (Corning) emulsion containing 100 μg Cre61–71 peptide (RKWFPAEPEDV, GenScript).
Thymic and Splenic B cell Isolation.
For isolation of thymic and splenic B cells, the tissue was first injected with 500 μL HBSS (with Ca2+,Mg2+) supplemented with Collagenase D (1 mg/mL, Roche,) 2% FCS and 10 mM HEPES, then finely minced in a total volume of 1 mL Collagense D solution and incubated for 30 min at 37 °C. Thymi were washed with HBSS (without Ca2+,Mg2+) supplemented with 2% FCS and 5 mM EDTA. Thymus suspensions were resuspended in PBS containing 2% FCS and stained with Fc block (2.4G2) for 15 min at 4 °C, washed, and then used for experiments.
Thymic B Cell Enrichment and RNA Isolation.
Processed thymic B cells were stained with biotinylated anti-CD4 and anti-CD8a antibodies, and thymocytes were depleted using anti-Biotin microbeads (Miltenyi Biotec) according to the manufacturer’s instructions. CD4/CD8a− thymocytes were surface stained with indicated antibodies, and CD19+B220+ IgM−IgD− were isolated using a FACSAria (BD Biosciences). The RNeasy Micro Kit (Qiagen) was used to isolate RNA obtained from each sample per the manufacturer’s instructions.
Bone Marrow Chimeras.
Bone marrow was isolated from the tibia and femur of mice. Bone marrow was isolated as previously described (56). Bone marrow chimera mice were generated by reconstituting lethally irradiated (1,000 rad) mice with 1 × 107 T cell–depleted donor bone marrow cells. Irradiated mice were given neomycin and polymyxin B supplemented water for at least 2 wk following irradiation and bone marrow transplantation. Chimeras were analyzed a minimum of 6 wk after reconstitution.
BrdU Labeling.
BrdU (Sigma-Aldrich) was reconstituted in sterile PBS and either injected intraperitoneal for a total dose of 2 mg or added to drinking water with 2% sucrose at a final concentration of 0.8 mg/mL. For BrdU staining, previously processed cell suspensions were surface stained as described above, followed by fixation and permeabilization using the BD Cytofix/Cytoperm kit per the manufacturer’s instructions. Following processing, cells were further permeabilized with BD Cytoperm buffer with 10% DMSO for 10 min at 4 °C, followed by two washes with BD Cytoperm buffer. Samples were then incubated with DNase (30 μg per 1 × 107 cells) diluted in PBS for 60 min at 37 °C followed by one wash in BD Cytoperm buffer. Cells were stained with anti-BrdU antibody for 20 min at 25 °C, washed in BD Cytoperm buffer analyzed by flow cytometry.
Tetramer Enrichment.
Secondary lymphoid organs (spleen and lymph nodes-axillary, brachial, inguinal, cervical, mesenteric, para-aortic) were isolated from mice and were pressed through a 70-μm nylon screen to generate a single-cell suspension. Single-cell suspensions were stained for 1 h at room temperature with allophycocyanin- or phycoerythrin-conjugated tetramers. Samples underwent enrichment for tetramer-binding cells as previously described (57). Cre61–71:I-Ab APC and PE tetramers were a generous gift from James Moon.
Cell Isolation and Flow Cytometry.
Thymi, spleen and Peyer’s patches (PPs) were isolated from mice. Spleen and thymi single-cell suspensions were generated as described above. PPs were pressed through a 70-μm nylon screen to generate a single-cell suspension. Single-cell suspensions were stained for 30 min at 4 °C with the indicated antibodies. Staining for CD40L was performed as previously described (58). For intracellular Ig H + L staining, the BD Cytofix/Cytoperm kit was used as per the manufacturer’s instructions. For intranuclear transcription factor staining, the Tonbo Foxp3 / Transcription Factor Staining Buffer Kit was used as per the manufacturer’s instructions. Samples were acquired with a BD LSRII or Fortessa X-20 (BD Biosciences) and analyzed with FlowJo version 10.8.1 (FlowJo LLC).
Intrathymic Transfers.
Spleens were isolated and pressed through a 70-μm nylon screen to generate a single-cell suspension. B cells were isolated using the EasySep Mouse B cell kit (StemCell Technologies) per the manufacturer’s protocols. CTV or CFSE labeling was performed following the manufacturer’s protocols. 5 × 106 purified B cells were injected into each lobe of the thymus using ultrasound guidance as previously described (59).
RT-qPCR.
From isolated RNA, cDNA was generated using the SuperScript III First-Strand Synthesis SuperMix for qRT-PCR (Thermo Fisher Scientific) as per the manufacturer’s protocol. RT-PCR was performed using Fast SYBR Green Master Mix (Roche) on the ABI PRISM 7900HT sequence detection system (Applied Bioscience). Gapdh was used for normalization of Ct values.
Immunofluorescence.
Thymi embedded in optimal cutting temperature compound (Sakura Finetek) were sectioned (7-μm slices) at −20 °C, followed by fixation and permeabilization in 100% acetone for 20 min at 4 °C. Acetone fixed sections were then blocked for 60 min at room temperature with 5% bovine serum albumin (BSA) and Fc block (clone 2.4G2). Following washing, sections were stained in 0.5% BSA and 0.1% Tween-20 (Sigma-Aldrich) overnight at 4 °C. Stained sections were washed, stained with DAPI, and mounted using Prolong antifade mounting medium (Life Technologies). Images of sections were acquired with a Leica DM6000B epifluorescent microscope
RNA Sequencing.
RNA sequencing was performed by the University of Minnesota Genomics Center. Total RNA was quantified using a RiboGreen assay. RNA quality was assessed via Agilent BioAnalyzer (Agilent Biotechnologies). Library creation was performed using the SMARTer Stranded Total RNA Pico Mammalian v2 kit (Takara Bio), according to the manufacturer’s instruction. Sequencing was performed on a NextSeq550 using paired-end 75-base chemistry at the University of Minnesota Genomics Center.
RNA Sequencing Analysis.
Reads were analyzed using the CHURP v0.2.1 pipeline developed by the University of Minnesota Supercomputing Institute (60). For the GSE152453 dataset (National Center for Biotechnology Information Gene Expression Omnibus), raw counts were analyzed using edgeR v3.24.3. GSEA was performed using Limma to generate a preranked list based on t-value and then analyzed using the GSEA-Broad Institute website application (https://www.gsea-msigdb.org/gsea/index.jsp). Heat maps of log2(CPM+1) normalized expression values were generated using the Morpheus website (https://software.broadinstitute.org/morpheus/). RNA-Seq data are available from the National Center for Biotechnology Information Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/) under accession GSE220104.
scRNA-seq and Analysis.
scRNA-seq experiments were performed as previously described (61). Briefly, MHC-II-expressing thymic cells were FACS sorted and captured using the 10× Genomics 3′ Single Cell V3 chemistry platform and were sequenced on a NovaSeq instrument. CellRanger (v 4.0.0) was used to process raw sequencing data, followed by the “count” function used to obtain gene (mRNA) and protein (HTO) count data for all cells. The Seurat R package (v3.2.2, R version 4.0.0) was used for analysis. Highly variable features (n = 2,000) from log normalized mRNA count data were identified by “FindVariableFeatures” function from the Seurat package, and HTO count data were added into the dataset, followed by centered-log ratio normalization. Doublet cells and cells derived from OT-I mice (by HTO) were identified using “HTODemux” and were removed from analysis. Principal components analysis and generation of a two-dimensional representation of data were generated using “RunPCA” and “RunUMAP,” respectively. Cells were clustered using the “FindNeighbors” and “FindClusters” Seurat functions. For the “FindClusters” function, different resolution values were tested and the resolution value of 0.1, which identified 14 clusters, was empirically chosen. The cluster representing B cells was selected, followed by selection of C57BL/6 and Ifnar1−/−/Ifnlr1−/−-derived cells (by HTO) and subset as a new object. This new object was reclustered as described above (resolution of 0.1). Differentially expressed genes from the WT B cells were identified by a Wilcoxon Rank Sum test implemented in the “FindMarkers” function in Seurat. For scoring of the expression of multiple genes of single cells, the Hallmark Interferon Alpha Response gene set was used as features to generate a new module score (“AddModuleScore” in Seurat) and plotted. Other MHCII-expressing thymic cells will be analyzed and reported elsewhere. scRNA-seq data generated are available from the National Center for Biotechnology Information Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/) under accession GSE192716.
Tcra RNA Sequencing.
Tcra sequencing was performed as previously described (2). Briefly, conventional CD4 single-positive T cells (CD8−CD4+CD25−GITR−) and Treg cells (CD8−CD4+CD25+GITR+) were FACS sorted from 6 to 12-wk-old AicdaCre/Cre or WT Tcra+/− TclibTg Foxp3eGFP thymi. Cells were sorted into RNAprotect Cell Reagent (Qiagen) and bulk RNA-seq of the Trav locus was performed using the RepSeq service from iRepertoire amplify all expressed Tcra transcripts. Samples were sequenced to a depth of five reads per cell. TCRs were analyzed based on the predicted amino acid sequence of the CDR3 region regardless of V gene usage and filtered to select for high confidence, recurrently expressed CDR3 clones. In brief, reads were normalized to counts per million (CPM), and data were filtered to include only CDR3 clones that were present at or above 10 CPM reads mapped in at least n-1 of biological replicates of at least one sample type. Analysis was performed using R (v. 4.0.5), including packages EdgeR, ggplot2 and pheatmap.
BCR N-Nucleotide Addition Analysis.
FASTQ files from 3H9 thymic and splenic B cells sequenced for Igk were downloaded from the Gene Expression Omnibus (GEO, GSE85366) and processed using IMGT/HighV-QUEST (62–65). Calculated Junction data were parsed using R (v. 4.0.5), and the frequency of productive Igk with no N-nucleotide additions was calculated.
Statistical Analysis.
Statistical analyses were performed using Prism 8 (GraphPad). For comparison of two datasets, two-tailed unpaired Student’s t tests were used. For comparison of three or more datasets, ordinary one-way ANOVA with Sidak’s multiple comparison tests or two-way ANOVA with Tukey’s multiple comparisons test was used. P-values less than or equal to 0.05 were considered significant. Sample size, experimental replicates, and additional details are provided in the figure legends.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
We thank J. Ding and L. Qian for technical assistance, J. Motl from the University Flow Cytometry Resource for cell sorting, J. Moon for providing tetramer reagents, the University of Minnesota Genomics Center for assistance with RNA-seq, scRNA-seq and TCR analysis and the University of Minnesota Research Animal Resources for animal husbandry. The scRNA seq data reported here were previously described in the thesis work of O.C.S. Matous Voboril is a Cancer Research Institute Irvington Fellow supported by the Cancer Research Institute (CRI Award # CRI4536). This project was supported by the NIH (grant nos. R37 AI39560 and P01 AI35296 to K.A.H., T32 AI007313 to K.M.A. and E.R.B., and F30 AI131483 and T32 GM008244 to E.R.B.).
Author contributions
R.J.M., E.R.B., K.M.A., M.V., O.C.S., and K.A.H. designed research; R.J.M., E.R.B., Y.W., K.M.A., M.V., T.M., and O.C.S. performed research; S.V.K. contributed new reagents/analytic tools; R.J.M., E.R.B., K.M.A., M.V., O.C.S., and C.H.O. analyzed data; and R.J.M. and K.A.H. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
Reviewers: E.A.H., The Ohio State University; and R.P., University of Colorado Anschutz Medical Campus.
Data, Materials, and Software Availability
RNA sequencing data have been deposited in Gene Expression Omnibus under accession numbers GSE220104 and GSE192716. All study data are included in the article and/or SI Appendix. A list of all materials used is included in SI Appendix, Table S1.
Supporting Information
References
- 1.Hogquist K. A., Baldwin T. A., Jameson S. C., Central tolerance: Learning self-control in the thymus. Nat. Rev. Immunol. 5, 772–782 (2005). [DOI] [PubMed] [Google Scholar]
- 2.Breed E. R., et al. , Type 2 cytokines in the thymus activate Sirpα+ dendritic cells to promote clonal deletion. Nat. Immunol. 23, 1042–1051 (2022), 10.1038/s41590-022-01218-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Breed E. R., Lee S. T., Hogquist K. A., Directing T cell fate: How thymic antigen presenting cells coordinate thymocyte selection. Semin. Cell Dev. Biol. 84, 2–10 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cepeda S., et al. , Age-associated decline in thymic B cell expression of aire and aire-dependent self-antigens. Cell Rep. 22, 1276–1287 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yamano T., et al. , Thymic B cells are licensed to present self antigens for central T cell tolerance induction. Immunity 42, 1048–1061 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Perera J., et al. , Self-antigen-driven thymic B cell class switching promotes T cell central tolerance. Cell Rep. 17, 387–398 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Walters S. N., Webster K. E., Daley S., Grey S. T., A role for Intrathymic B cells in the Generation of natural regulatory T cells. J. I. 193, 170–176 (2014). [DOI] [PubMed] [Google Scholar]
- 8.Lu F.-T., et al. , Thymic B cells promote thymus-derived regulatory T cell development and proliferation. J. Autoimmun. 61, 62–72 (2015). [DOI] [PubMed] [Google Scholar]
- 9.Perera J., Meng L., Meng F., Huang H., Autoreactive thymic B cells are efficient antigen-presenting cells of cognate self-antigens for T cell negative selection. Proc. Natl. Acad. Sci. U.S.A. 110, 17011–17016 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marnik E. A., et al. , Precocious interleukin 21 expression in naive mice identifies a natural helper cell population in autoimmune disease. Cell Rep. 21, 208–221 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tangye S. G., Ma C. S., Regulation of the germinal center and humoral immunity by interleukin-21. J. Exp. Med. 217, e20191638 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Victora G. D., Nussenzweig M. C., Germinal centers. Annu. Rev. Immunol. 30, 429–457 (2012). [DOI] [PubMed] [Google Scholar]
- 13.Chen J. S., et al. , High-affinity, neutralizing antibodies to SARS-CoV-2 can be made without T follicular helper cells. Sci. Immunol. 7, eabl5652 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roco J. A., et al. , Class-switch recombination occurs infrequently in germinal centers. Immunity 51, 337–350.e7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Taylor J. J., Pape K. A., Jenkins M. K., A germinal center–independent pathway generates unswitched memory B cells early in the primary response. J. Exp. Med. 209, 597–606 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cordero H., et al. , Intrathymic differentiation of natural antibody-producing plasma cells in human neonates. Nat. Commun. 12, 5761 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barwick B. G., Scharer C. D., Bally A. P. R., Boss J. M., Plasma cell differentiation is coupled to division-dependent DNA hypomethylation and gene regulation. Nat. Immunol. 17, 1216–1225 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hodgkin P. D., Lee J. H., Lyons A. B., B cell differentiation and isotype switching is related to division cycle number. J. Exp. Med. 184, 277–281 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Andreu-Sánchez J. L., et al. , Ontogenic characterization of thymic B lymphocytes. Analysis in different mouse strains. Eur. J. Immunol. 20, 1767–1773 (1990). [DOI] [PubMed] [Google Scholar]
- 20.Dong M., et al. , Alterations in the thymic selection threshold skew the self-reactivity of the TCR repertoire in neonates. J. Immunol. 199, 965–973 (2017). [DOI] [PubMed] [Google Scholar]
- 21.Gilfillan S., Dierich A., Lemeur M., Benoist C., Mathis D., Mice lacking TdT: Mature animals with an immature lymphocyte repertoire. Science 261, 1175–1178 (1993). [DOI] [PubMed] [Google Scholar]
- 22.Fujihara C., et al. , T Cell–B cell thymic cross-talk: Maintenance and function of thymic B cells requires cognate CD40–CD40 ligand interaction. J. Immunol. 193, 5534–5544 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kwon D., et al. , Homeostatic serum IgE is secreted by plasma cells in the thymus and enhances mast cell survival. Nat. Commun. 13, 1418 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baumgarth N., Tung J. W., Herzenberg L. A., Inherent specificities in natural antibodies: A key to immune defense against pathogen invasion. Springer Semin. Immun. 26, 347–362 (2005). [DOI] [PubMed] [Google Scholar]
- 25.Montecino-Rodriguez E., Leathers H., Dorshkind K., Identification of a B-1 B cell–specified progenitor. Nat. Immunol. 7, 293–301 (2006). [DOI] [PubMed] [Google Scholar]
- 26.Vinuesa C. G., Linterman M. A., Yu D., MacLennan I. C. M., Follicular helper T Cells. Annu. Rev. Immunol. 34, 335–368 (2016). [DOI] [PubMed] [Google Scholar]
- 27.Klein U., et al. , Transcriptional analysis of the B cell germinal center reaction. Proc. Natl. Acad. Sci. U.S.A. 100, 2639–2644 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Choi Y. S., et al. , ICOS receptor instructs T follicular helper cell versus effector cell differentiation via induction of the transcriptional repressor Bcl6. Immunity 34, 932–946 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kiefer K., Oropallo M. A., Cancro M. P., Marshak-Rothstein A., Role of type I interferons in the activation of autoreactive B cells. Immunol. Cell Biol. 90, 498–504 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lienenklaus S., et al. , Novel reporter mouse reveals constitutive and inflammatory expression of IFN-β in vivo. J. Immunol. 183, 3229–3236 (2009). [DOI] [PubMed] [Google Scholar]
- 31.Benhammadi M., et al. , IFN-λ enhances constitutive expression of MHC class I molecules on thymic epithelial cells. J. Immunol. 205, 1268–1280 (2020). [DOI] [PubMed] [Google Scholar]
- 32.Uccellini M. B., García-Sastre A., ISRE-reporter mouse reveals high basal and induced type I IFN responses in inflammatory monocytes. Cell Rep. 25, 2784–2796.e3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Durbin J. E., Hackenmiller R., Simon M. C., Levy D. E., Targeted disruption of the mouse Stat1 gene results in compromised innate immunity to viral disease. Cell 84, 443–450 (1996). [DOI] [PubMed] [Google Scholar]
- 34.Huang S., et al. , Immune response in mice that lack the interferon-γ receptor. Science 259, 1742–1745 (1993). [DOI] [PubMed] [Google Scholar]
- 35.Lin J.-D., et al. , Distinct roles of type I and type III interferons in intestinal immunity to homologous and heterologous rotavirus infections. PLoS Pathog. 12, e1005600 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ferrero I., et al. , Functional and phenotypic analysis of thymic B cells: Role in the induction of T cell negative selection. Eur. J. Immunol. 29, 1598–1609 (1999). [DOI] [PubMed] [Google Scholar]
- 37.Frommer F., Waisman A., B cells participate in thymic negative selection of murine auto-reactive CD4+ T cells. PLoS One 5, e15372 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Legoux F. P., et al. , CD4 + T cell tolerance to tissue-restricted self antigens is mediated by antigen-specific regulatory T cells rather than deletion. Immunity 43, 896–908 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Owen D. L., et al. , Thymic regulatory T cells arise via two distinct developmental programs. Nat. Immunol. 20, 195–205 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hsieh C.-S., et al. , Recognition of the peripheral self by naturally arising CD25+ CD4+ T cell receptors. Immunity 21, 267–277 (2004). [DOI] [PubMed] [Google Scholar]
- 41.Wong P., Goldrath A. W., Rudensky A. Y., Competition for specific intrathymic ligands limits positive selection in a TCR Transgenic model of CD4 + T cell development. J. Immunol. 164, 6252–6259 (2000). [DOI] [PubMed] [Google Scholar]
- 42.Durbin R. K., Kotenko S. V., Durbin J. E., Interferon induction and function at the mucosal surface. Immunol. Rev. 255, 25–39 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sommereyns C., Paul S., Staeheli P., Michiels T., IFN-lambda (IFN-λ) is expressed in a tissue-dependent fashion and primarily acts on epithelial cells in vivo. PLoS Pathog. 4, e1000017 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ye L., et al. , Interferon-λ enhances adaptive mucosal immunity by boosting release of thymic stromal lymphopoietin. Nat. Immunol. 20, 593–601 (2019). [DOI] [PubMed] [Google Scholar]
- 45.Hahn W. O., Pepper M., Liles W. C., B cell intrinsic expression of IFNλ receptor suppresses the acute humoral immune response to experimental blood-stage malaria. Virulence 11, 594–606 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Syedbasha M., et al. , Interferon-λ enhances the differentiation of naive B cells into plasmablasts via the mTORC1 pathway. Cell Rep. 33, 108211 (2020). [DOI] [PubMed] [Google Scholar]
- 47.Miller C. N., et al. , Thymic tuft cells promote an IL-4-enriched medulla and shape thymocyte development. Nature 559, 627–631 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yano M., et al. , Aire controls the differentiation program of thymic epithelial cells in the medulla for the establishment of self-tolerance. J. Exp. Med. 205, 2827–2838 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rudensky A. Y., Mazel S. M., Yurin V. L., Presentation of endogenous immunoglobulin determinant to immunoglobulin-recognizing T cell clones by the thymic cells. Eur. J. Immunol. 20, 2235–2239 (1990). [DOI] [PubMed] [Google Scholar]
- 50.Adler L. N., et al. , The other function: Class II-restricted antigen presentation by B cells. Front. Immunol. 8, 319 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huszthy P. C., et al. , B cell receptor ligation induces display of V-region peptides on MHC class II molecules to T cells. Proc. Natl. Acad. Sci. U.S.A. 116, 25850–25859 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gray D. H. D., et al. , Developmental kinetics, turnover, and stimulatory capacity of thymic epithelial cells. Blood 108, 3777–3785 (2006). [DOI] [PubMed] [Google Scholar]
- 53.Hirakawa M., et al. , Fundamental parameters of the developing thymic epithelium in the mouse. Sci. Rep. 8, 11095 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Deng J., Wei Y., Fonseca V. R., Graca L., Yu D., T follicular helper cells and T follicular regulatory cells in rheumatic diseases. Nat. Rev. Rheumatol. 15, 475–490 (2019). [DOI] [PubMed] [Google Scholar]
- 55.Vinuesa C. G., Sanz I., Cook M. C., Dysregulation of germinal centres in autoimmune disease. Nat. Rev. Immunol. 9, 845–857 (2009). [DOI] [PubMed] [Google Scholar]
- 56.Amend S. R., Valkenburg K. C., Pienta K. J., Murine hind limb long bone dissection and bone marrow isolation. J. Vis. Exp. 14, 53936 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Moon J. J., et al. , Tracking epitope-specific T cells. Nat. Protoc. 4, 565–581 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Koguchi Y., Thauland T. J., Slifka M. K., Parker D. C., Preformed CD40 ligand exists in secretory lysosomes in effector and memory CD4+ T cells and is quickly expressed on the cell surface in an antigen-specific manner. Blood 110, 2520–2527 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang H., et al. , Ultrasound guided intra-thymic injection to track recent thymic emigrants and investigate T cell development. Bio Protoc. 8, e3107 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Baller J., Kono T., Herman A., Zhang Y., “CHURP: A lightweight CLI framework to enable novice users to analyze sequencing datasets in parallel” inProceedings of the Practice and Experience in Advanced Research Computing on Rise of the Machines (Learning) (ACM, 2019), pp. 1–5. [Google Scholar]
- 61.Barrero O. C. S., “Thymic interferons and protein O-GlcNAcylation in regulatory T cells: two tales of T cell tolerance,” PhD thesis, University of Minnesota, University Digital Conservancy, MN (2021).
- 62.Alamyar E., Duroux P., Lefranc M.-P., Giudicelli V., IMGT® tools for the iucleotide analysis of immunoglobulin (IG) and T cell receptor (TR) V-(D)-J repertoires, polymorphisms, and IG mutations: IMGT/V-QUEST and IMGT/HighV-QUEST for NGS in Immunogenetics: Methods and Applications in Clinical Practice, Christiansen F. T., Tait B. D., Eds. (Humana Press, 2012), pp. 569–604. [DOI] [PubMed] [Google Scholar]
- 63.Giudicelli V., Chaume D., Lefranc M.-P., IMGT/GENE-DB: A comprehensive database for human and mouse immunoglobulin and T cell receptor genes. Nucleic Acids Res. 33, D256–D261 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Giudicelli V., et al. , From IMGT-ONTOLOGY to IMGT/HighVQUEST for NGS Immunoglobulin (IG) and T cell receptor (TR) repertoires in autoimmune and infectious diseases. Autoimmun. Infec. Dis. 1 (2015), 10.16966/2470-1025.103. [DOI] [Google Scholar]
- 65.Alamyar E., Giudicelli V., Li S., Duroux P., Lefranc M.-P., Imgt/Highv-quest: The Imgt® web portal For immunoglobulin (Ig) or antibody and T cell receptor (Tr) analysis from Ngs high throughput and deep sequencing. Immunome Res. 8 (2012), 10.4172/1745-7580.1000056. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
RNA sequencing data have been deposited in Gene Expression Omnibus under accession numbers GSE220104 and GSE192716. All study data are included in the article and/or SI Appendix. A list of all materials used is included in SI Appendix, Table S1.