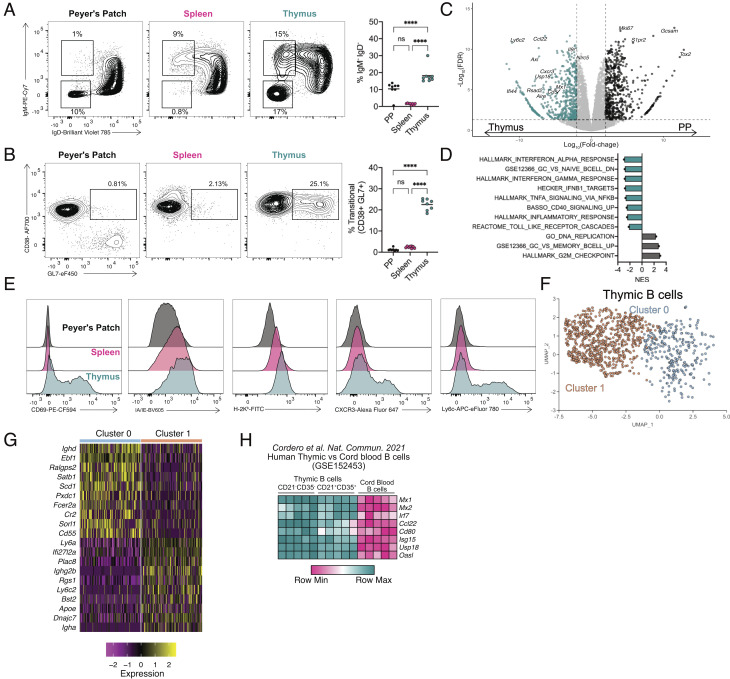
Fig. 1.
Class-switched thymic B cells exhibit a unique phenotype. (A) Flow cytometry of B cells from Peyer’s patch (Left), spleen (Middle), and thymus (Right) showing IgM and IgD expression, with number adjacent to outlined areas identifying proportion of isotype class-switched (IgM−IgD−) and activated IgM+IgDlo B cells. Graph on right shows quantification of isotype class-switched B cells (IgM−IgD−). (B) Flow cytometry of total B cells from the Peyer’s patch (Left), spleen (Middle), and thymus (Right) showing CD38 and GL7 expression with number adjacent to outlined areas identifying proportion of activated memory B cell precursors. Graph on Right shows quantification of activated memory B cell precursors (CD38+GL7+). (C) Volcano plot representation of RNA-Seq data comparing isotype class-switched thymic and PP B cells showing Log2 fold-change by -Log10 FDR. Significantly up-regulated genes (FDR <= 0.05) are identified in the thymus (blue, FC < −2) and PP (black, FC > 2). (D) Gene set enrichment analysis normalized enhancement score (NES) for ranked RNA-Seq data. Negative NES are pathways overrepresented in thymic B cells; positive NES pathways overrepresented in PP B cells. (E) Flow cytometry histograms for CD69, I-A/I-E, H2-Kb, CXCR3, and Ly6C expression of B cells in the PP (black, Top), spleen (pink, Middle), and thymus (blue, Bottom). (F) Uniform Manifold Approximation and Projection (UMAP) plot for thymic B cells. (G) Heatmap representation of top 10 differentially expressed genes in subsampled thymic B cells from cluster 0 and 1. (H) Heatmap representation of log2-transformed counts per-million (CPM) of individual ISG mRNA in CD21−CD35− and CD21+CD35+ thymic B cells compared with cord blood B cells. Each symbol (A and B) represents an individual mouse; small horizontal lines indicate the group mean. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, and ****P ≤ 0.0001 (A and B One-way ANOVA with Tukey’s multiple comparisons test).