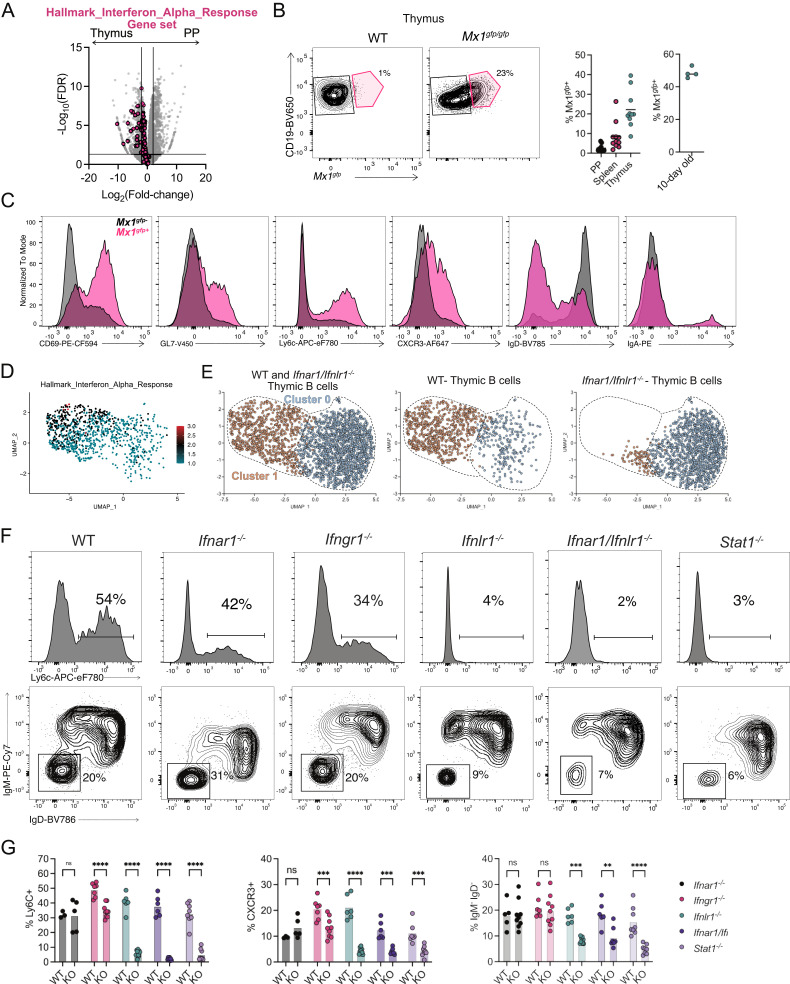
Fig. 4.
Type III IFN drives thymic B cell licensing. (A) Volcano plot representation of RNA-Seq data comparing isotype class-switched thymic and PP B cells (gray) with overlaid “Hallmark_Interferon_Alpha_Response” gene set (pink). (B) Flow cytometry of thymic B cells showing CD19 and Mx1gfpin WT and Mx1gfp/gfp mice with number adjacent to outlined areas identifying proportion of Mx1gfp+ thymic B cells. Graphs on right showing quantification of Mx1gfp+ PP, splenic and thymic B cells in adult mice, and thymic B cells from 10-d-old mice (C) Flow cytometry histogram overlays of Mx1gfp+ (pink) and Mx1gfp− (black) thymic B cells showing expression of CD69, GL7, Ly6C, CXCR3, IgD, and IgA. (D) UMAP plot of thymic B cells with module score identifying cells expressing genes identified in the “Hallmark_Interferon_Alpha_Response” gene set. (E) UMAP plots of thymic B cells derived from WT and Infar1−/−/Inflr1−/− hosts identified by HTO. (F) Flow cytometry histograms showing Ly6C expression (Top) or isotype class-switched (IgM−IgD−) thymic B cells derived from WT, Ifnar1−/−, Ifngr1−/−, Ifnlr1−/−, Ifnar1−/−/Ifnlr1−/−, and Stat1−/− mice. Numbers adjacent to outlined areas identifying the proportion of cells within the gate. (G) Quantification of Ly6C+, CXCR3+, and isotype class-switched (IgM−IgD−) thymic B cells from WT, Ifnar1−/−, Ifngr1−/−, Ifnlr1−/−, Ifnar1−/−/Ifnlr1−/,− and Stat1−/− mice. Each symbol (B and G) represents an individual mouse; small horizontal lines indicate the group mean. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001 and ****P ≤ 0.0001 [(G) Two-way ANOVA with Sidak’s multiple comparisons test].