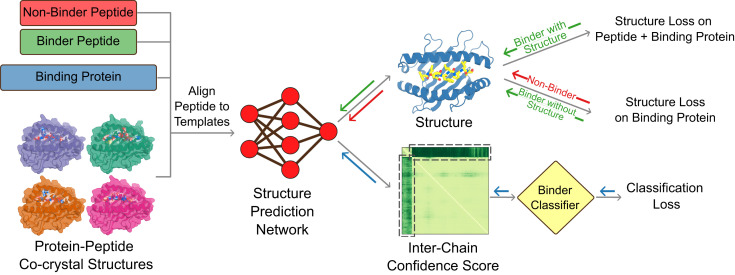
Fig. 1.
Extending structure prediction networks for binder classification. For fine-tuning the combined model to distinguish binding from non-binding peptides, inputs (Left) are sets of known binding (green) and non-binding (red) sequences, the sequence of the target protein(s), and the available co-crystal structures which provide templates for modeling the complex. The peptide sequence is positionally aligned to the peptides in the co-crystal structures, and the structure of the complex is predicted with AlphaFold. A logistic regression classifier converts the output PAE values into a normalized binder/non-binder score and prediction. During training, the combined model parameters were optimized to minimize the loss terms in the final column, and model weights were updated through gradient backpropagation as indicated by the solid arrows. The classification loss (cross-entropy) is computed on all training examples; the structure loss is computed over the entire complex for binding peptides with co-crystal structures and over the binding protein alone for binding peptides without co-crystal structures and for non-binding peptides.