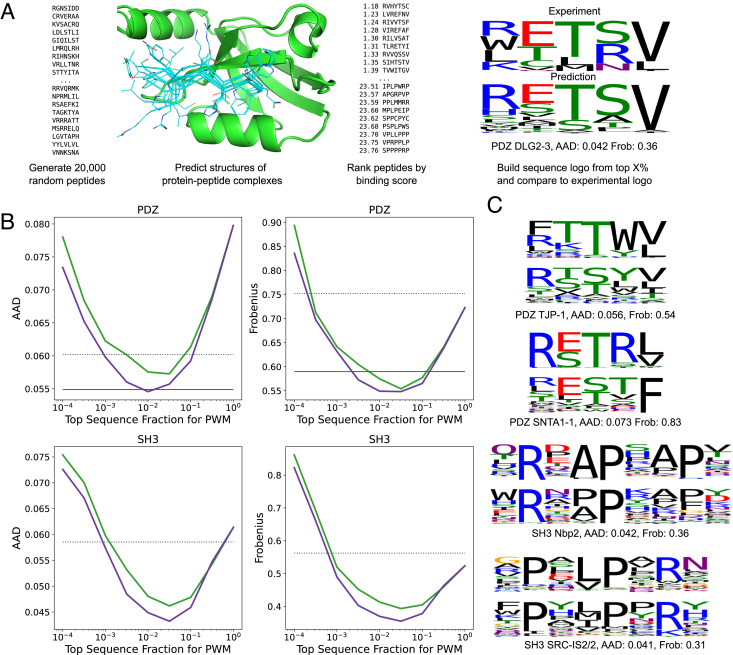
Fig. 4.
Accurate prediction of PDZ-peptide and SH3-peptide interactions using the combined structure prediction-classification network. (A) Prediction of position-weight matrices by random sequence generation followed by network evaluation. (B) Combined network fine-tuned on peptide-MHC interactions outperforms AlphaFold with default parameters in recapitulating experimental PWMs. AAD: average absolute difference in amino acid frequency; Frobenius: average Frobenius distance between PWM columns (lower is better for both; see Methods). AlphaFold (green) and fine-tuned AlphaFold (purple). The solid black lines in the PDZ panels indicate the performance of the structure-based method from ref. 20, which was optimized on PDZ domains. Dashed black lines indicate the average AAD and Frobenius scores of PWMs constructed from the sequences of the template PDB peptides used for modeling. (C) Examples of predicted and experimental sequence logos (see SI Appendix, Figs. S9 and S10 for a complete set of sequence logos).