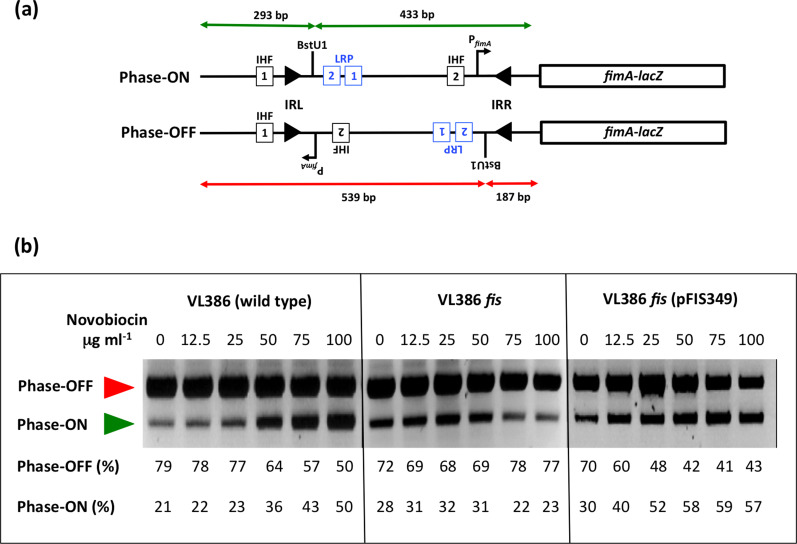
Fig. 1.
The phase OFF-to-phase ON bias of fimS genetic switch inversion in novobiocin-treated cultures is reversed in the absence of the Fis protein. (a) The fimS inversion assay. The fimS genetic element is amplified from the chromosome by PCR and the amplimers are cleaved with the BstUI restriction endonuclease (see Methods). The lengths of the cleavage products are summarized for the ON (green) and OFF (red) orientations of fimS, above and below the drawings, respectively. The angled arrow, labelled P fimA , shows the position and orientation of the transcriptional promoter of the fimA gene. In strain VL386 and its derivatives, the fimA gene is fused to a lacZ reporter gene, allowing phase ON and phase OFF bacterial colonies to be distinguished on MacConkey lactose indicator agar plates. Squares represent the binding sites for IHF (black) and Lrp (blue), respectively. The filled arrowheads represent the left (IRL) and right (IRR) 9 bp inverted repeats that flank fimS. The position of the BstUI restriction endonuclease recognition site between Lrp binding site LRP-2 and IRR is shown. Not to scale. (b) Electrophoresis of the fimS DNA fragments from the wild-type strain (VL386), its fis knockout derivative and the complemented fis mutant, following BstUI digestion of the PCR-amplified fimS genetic element. The red arrowhead indicates the 539 bp phase OFF diagnostic band and the green arrowhead shows the 433 bp diagnostic band. The cultures had been treated with novobiocin at the concentrations given above each gel lane. The intensities of the DNA bands corresponding to the ON and OFF orientations of fimS in each lane were determined by densitometry and are reported as percentages below the lane. The experiment was performed three times and typical data are presented.