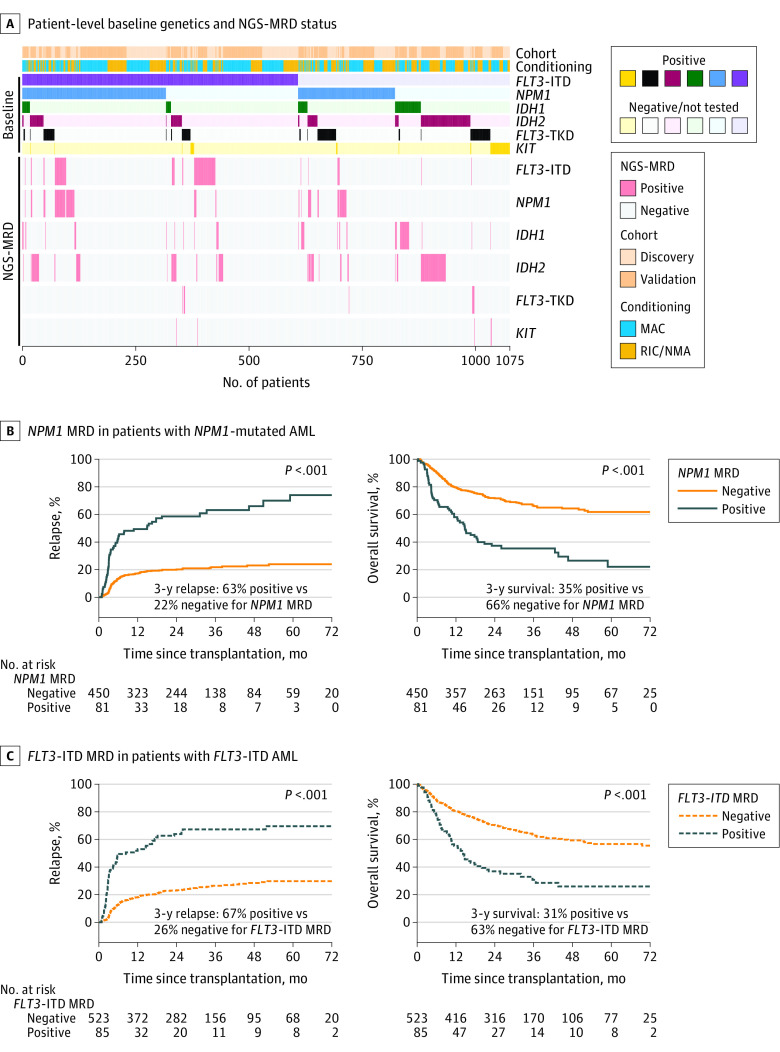
Figure 3. Detection and Impact of Residual Variants on Clinical Outcomes by Gene.
A, Heatmap illustrating for each patient (n = 1075) their cohort assignment, conditioning intensity, reported baseline variant status, and residual variant detection status by next-generation sequencing measurable residual disease (NGS-MRD) for each gene. Patients are sorted by the variants reported to the Center for International Blood and Marrow Transplant Research as being present at initial (baseline) diagnosis (positive or negative/not tested).
B, Patients with NPM1 mutated acute myeloid leukemia (AML) (n = 531) with residual NPM1 variants detectable by NGS prior to transplant (NPM1 MRD positive) had significantly higher rates of relapse and decreased overall survival than patients with no NPM1 variants detected (NPM1 MRD negative). The median time of observation was 29 months (IQR, 24-48 months) for patients who tested positive and 34 months (IQR, 24-50 months) for those who tested negative.
C, Patients with FLT3 internal tandem duplication (FLT3-ITD) AML (n = 608) with residual FLT3-ITD variants detectable by NGS prior to transplant (FLT3-ITD MRD positive) had significantly higher rates of relapse and decreased overall survival than patients with no FLT3-ITD detected (FLT3-ITD MRD negative). The median time of observation was 28 months (IQR, 24-61 months) in patients who tested positive and 35 months (IQR, 24-52 months) for those who tested negative.
MAC indicates myeloablative conditioning, NMA, nonmyeloablative conditioning; RIC, reduced-intensity conditioning; TKD, tyrosine kinase domain.