Figure 5. pH-dependent intra-protomer contacts govern ligand binding site availability.
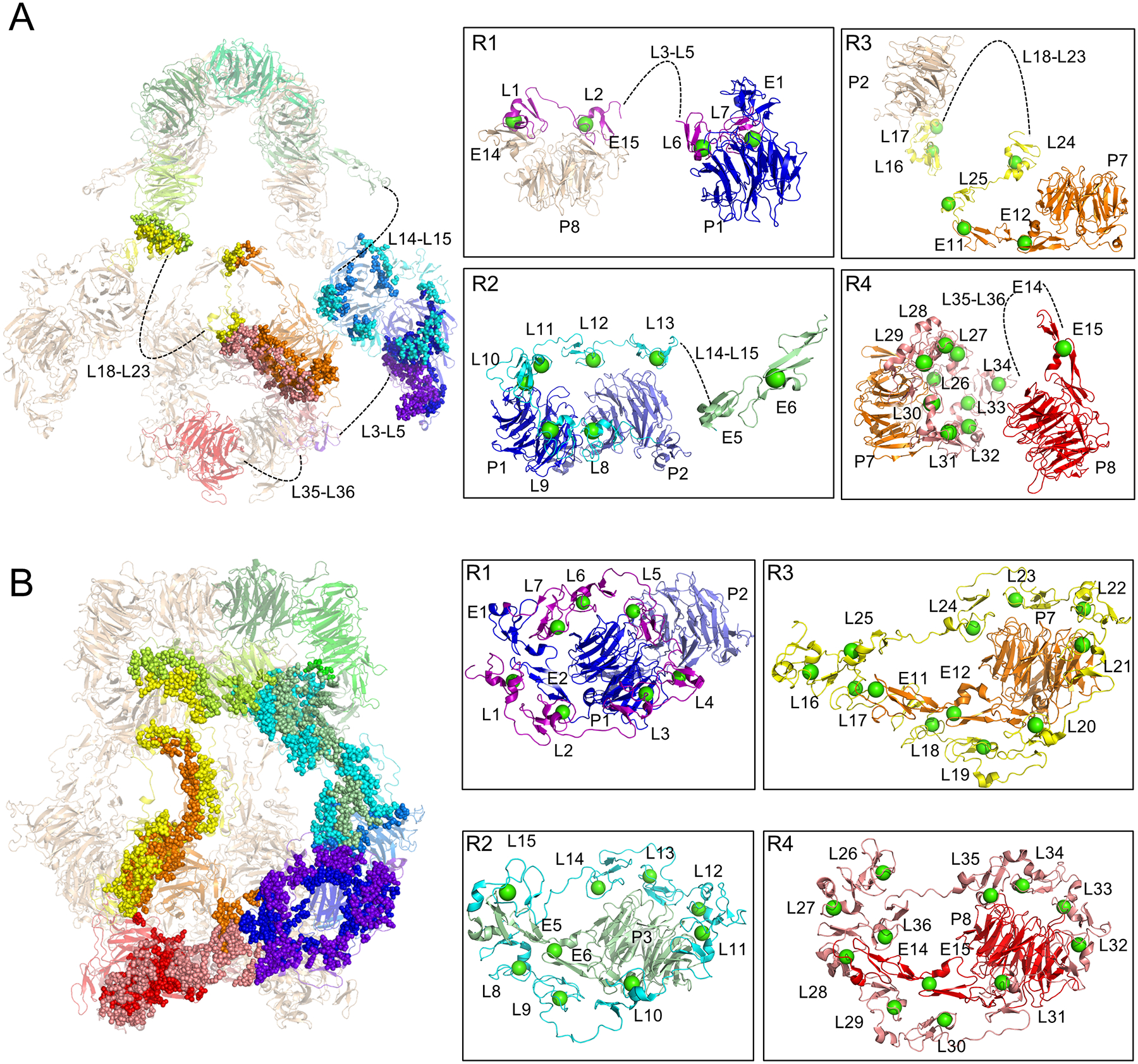
(A) Residues involved in intra-protomer contacts at pH 7.5 are rendered as spheres superimposed on a semi-transparent cartoon depiction of the LRP2 structure (left panel). Flexible ligand-binding repeats that could not be modeled are indicated with dashed lines and labeled. Residue level interactions are shown at right in boxes for each of the four ligand-binding domains. Ca2+ ions in the L repeats are rendered as green spheres. Individual ligand-binding repeats within the larger domains are labeled. Domains from one protomer are colored in rainbow, and from the other protomer in wheat.
(B) Residues involved in intra-protomer contacts at pH 5.2 are rendered as spheres superimposed on a transparent cartoon depiction of the structure (left panel).
See also Figure S4
