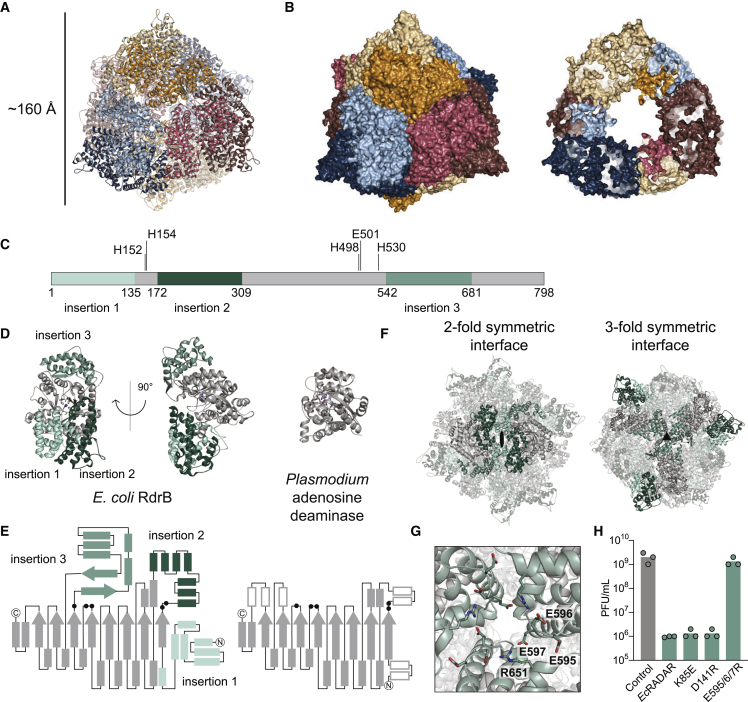
Figure 3.
Structure of RdrB reveals an adenosine deaminase dodecamer
(A) Cartoon representation of E. coli RdrB dodecamer, with each protomer in a different color.
(B) Surface representation of E. coli RdrB dodecamer (left), and slice through showing hollow center (right).
(C) Schematic representation showing insertions in RdrB relative to other adenosine deaminases.
(D and E) Structure (D) and schematic (E) representation of single RdrB monomer highlighting insertions in shades of green (left) compared with Plasmodium adenosine deaminase (PDB: 2PGF) (right). Active site residues are shown as black dots in the schematic (E).
(F) Insertions 1 and 2 form 2-fold symmetric dodecamer interface and insertion 3 forms the 3-fold symmetric dodecamer interface.
(G) Close-up view of 3-fold interface shown in (F) highlighting key residues, including those mutated to disrupt the interface in (H).
(H) Effect of point mutations suggested to disrupt the RdrB multimerization interfaces on the defensive activity of EcRADAR. Data represent PFU mL−1 of T2 phage infecting control cells, EcRADAR-expressing cells. Shown is the average of three replicates, with individual data points overlaid. The same control replicates are shown in Figures 1E and 4G for reference.