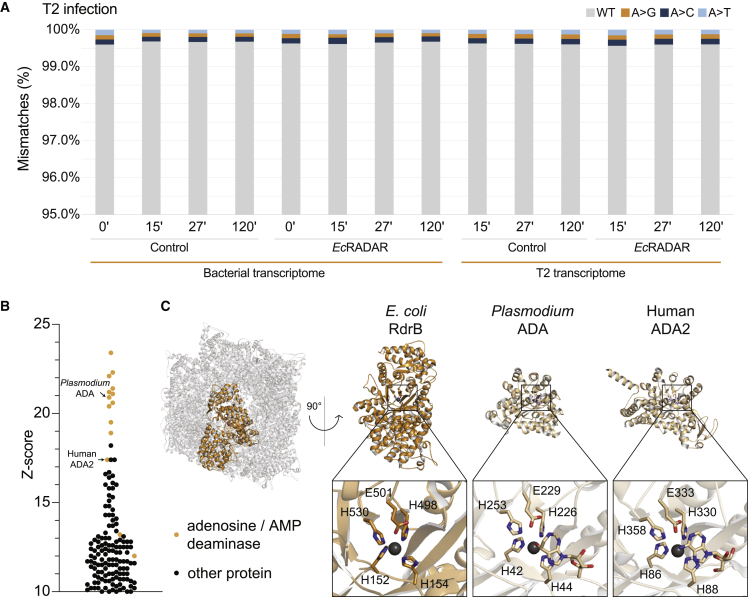
Figure 5.
Structural analysis of RdrB suggests targeting of nucleotide substrates
(A) Mismatches between sequenced RNA and genomic DNA in adenine positions. RNA, extracted from EcRADAR-expressing or control cells infected by phage T2, was sequenced and the resulting reads were aligned to the reference sequences of the E. coli host and the T2 phage genomes. Shown in the rate of mismatches in all expressed adenosine positions mapped to non-rRNA genes. x axis depicts the time from the onset of infection, with t = 0 reflecting uninfected cells; y axis is the observed rate of mismatches.
(B) DALI Z score of protein structures similar to RdrB, showing homology to adenosine and AMP deaminases (highlighted in yellow).
(C) Cartoon representation of RdrB dodecamer, highlighting a single monomer in yellow (left), and comparison to other adenosine deaminase active sites (right).