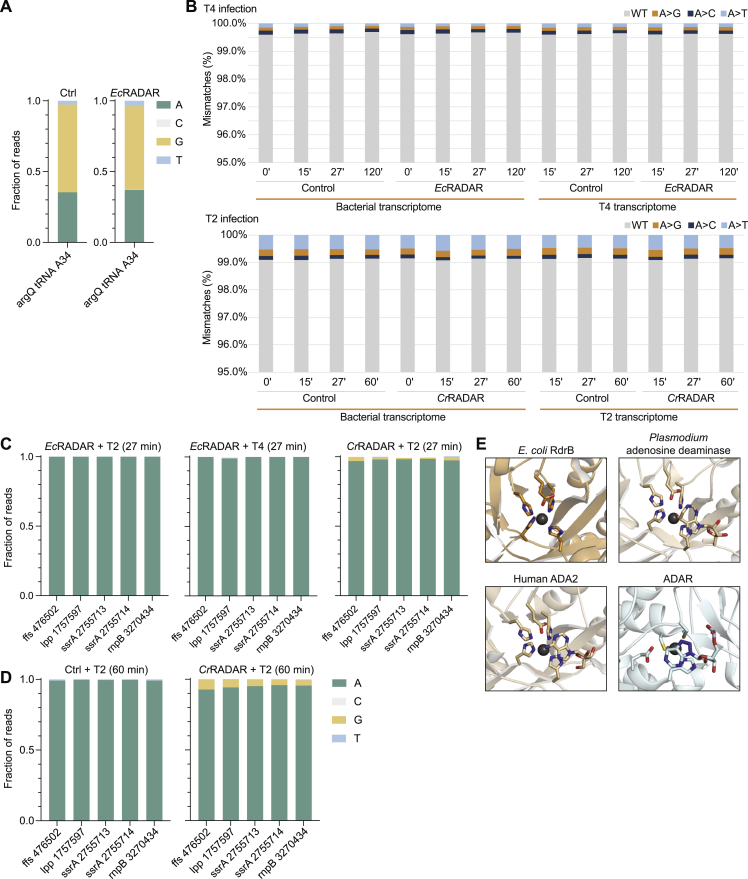
Figure S5.
Analysis of deaminase activity of RdrB, related to Figure 5
(A) Detection of A-to-G mutational signature caused by A-to-I editing of the arginine tRNA argQ, confirming the ability of the RNA-seq pipeline to detect editing in control and RADAR-containing uninfected cells.
(B) Mismatches between sequenced RNA and genomic DNA in adenine positions. RNA, extracted from EcRADAR and CrRADAR-expressing cells or control cells infected by phages T4 or T2 (respectively), was sequenced and the resulting reads were aligned to the reference sequences of the E. coli host and the respective phage genome. Shown in the rate of mismatches in all expressed adenosine positions mapped to non-rRNA genes. x axis depicts the time from the onset of infection, with t = 0 reflecting uninfected cells; y axis is the observed rate of mismatches.
(C) A-to-G mutational signature in T2- and T4-infected cells with or without EcRADAR and CrRADAR 27 min after infection, showing select genome locations that are abundantly expressed and were previously reported to contain an A-to-G signature.6
(D) A-to-G mutational signature in T2-infected cells with or without CrRADAR 60 min after infection, showing select genome locations that are abundantly expressed and were previously reported to contain an A-to-G signature.6
(E) Comparison of adenosine deaminase active sites of enzymes that modify monomeric substrates (RdrB, ADA, and ADA2) and ADAR, which modifies RNA substrates using a distinct active site and is part of the cytidine deaminase superfamily.