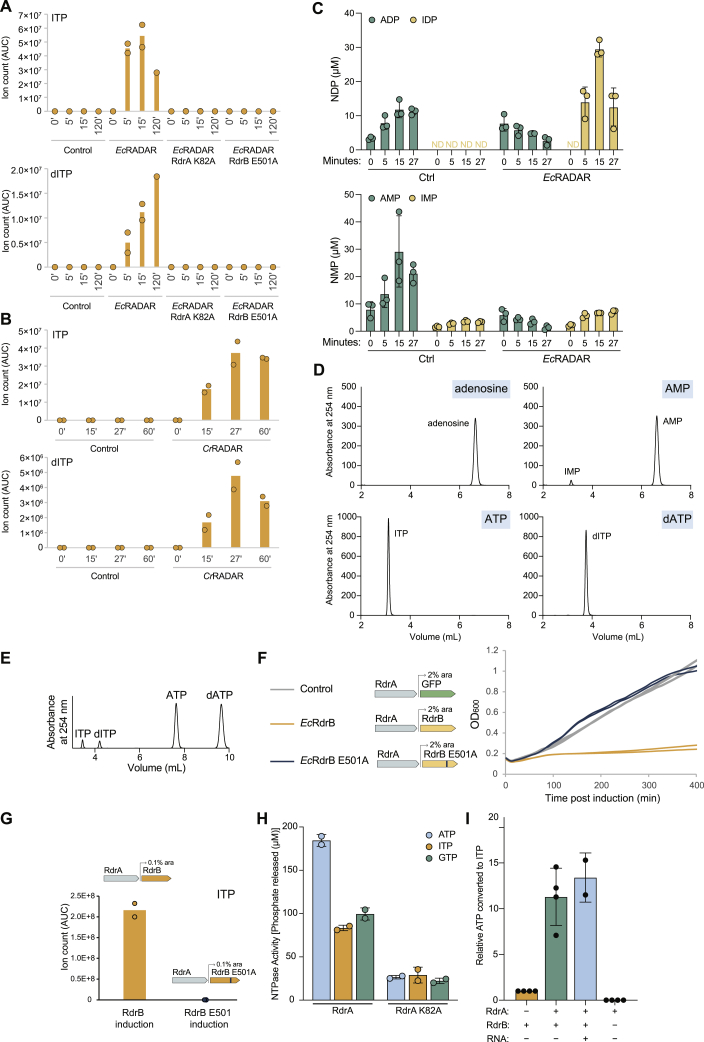
Figure S6.
RdrB converts (d)ATP to (d)ITP in vivo and in vitro, related to Figure 6
(A) Ion count (area under curve) of ITP or dITP (respectively) in lysates extracted from WT EcRADAR containing cells, as well as RADAR mutated in RdrA-K82A or RdrB-E501A, as measured by LC-MS/MS. The x axis represents min after infection, with zero representing non-infected cells. Cells were infected by phage T4 at an MOI of 2 at 37°C. Bar graphs represent the average of two biological replicates, with individual data points overlaid.
(B) Ion count (area under curve) of ITP or dITP (respectively) in lysates extracted from WT CrRADAR containing cells, or control cells, as measured by LC-MS/MS. The x axis represents min after infection, with zero representing non-infected cells. Cells were infected by phage T2 at an MOI of 2 at 37°C. Bar graphs represent the average of two biological replicates, with individual data points overlaid.
(C) Quantitative mass spectrometry of ADP, AMP, IDP, and IMP in lysates extracted from cells containing EcRADAR or control plasmid. Cells were infected with T4 phage at an MOI of 2 for indicated amount of time before harvesting. Bar graph shows the mean, with error bars representing standard deviation.
(D) Full HPLC traces of data summarized in Figure 6C.
(E) HPLC analysis of ATP and dATP co-incubated with RdrB, showing similar deamination of both substrates.
(F) Growth curves of cells expressing EcRdrB (orange), the mutant EcRdrB E501A (blue), and control cells expressing GFP (gray) in the presence of 2% arabinose without phage infection. Results of two experiments are presented as individual curves.
(G) Ion count (area under curve) of ITP in lysates extracted from EcRdrB and EcRdrB E501A expressing cells, as measured by LC-MS/MS. Cells were supplemented with 0.1% arabinose and incubated at 37°C for 100 min. Bar graphs represent the average of two biological replicates, with individual data points overlaid.
(H) Phosphate release assay measuring NTP hydrolysis by RdrA. Purified RdrA was incubated with indicated NTP, then phosphate released by hydrolysis was quantified using malachite green. Bar graph shows the mean, with error bars representing standard deviation.
(I) ATP-to-ITP conversion by RdrB either alone, in the presence of RdrA, or in the presence of RdrA and hairpin RNA. RdrA alone does not deaminate ATP to ITP. Bar graph shows the mean, with error bars representing standard deviation.