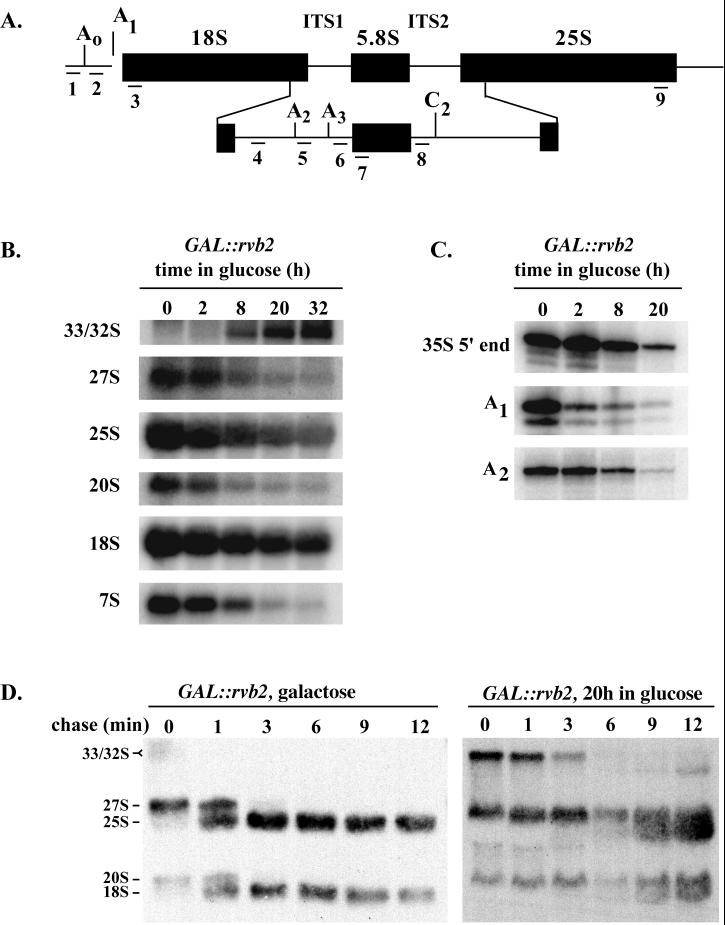
FIG. 4.
Rvb2p is required for pre-rRNA processing. (A) Structure of the 35S pre-rRNA, showing the positions of cleavage sites and regions complementary to oligonucleotide probes 1 to 8. ITS, internal transcribed spacer. (B) Steady-state levels of pre-rRNA species in cells depleted of Rvb2 mRNA for 0 to 32 h. Total RNA was extracted from cells at the times indicated following shift from galactose to glucose and subjected to Northern analysis. Loading was adjusted to obtain approximately equal quantities of tRNA. The oligonucleotides used to identify the various rRNA species are as follows: probes 3 and 9, mature 18S and 25S rRNAs, respectively; probe 4, 20S and 35S precursors; probe 8, 27S and 7S precursors. (C) Mapping of the 5′ ends of the 35S precursor and products of the A1 and A2 cleavage reactions. The 5′ ends were identified by primer extension analysis using as the template total RNA from cells depleted of Rvb2 mRNA as described for panel B. Oligonucleotides 1, 3, and 5 were extended to the 5′ end of 35S rRNA and the A1 and A2 cleavage sites, respectively. (D) Pulse-chase analysis of pre-rRNA processing in GAL::rvb2 cells incubated in galactose (left) or glucose medium for 20 h (right). Cells were labeled with [3H]methionine for 2.5 min and chased with cold methionine. Samples were removed at the times indicated, and total RNA was extracted and fractionated on an agarose-formaldehyde gel.