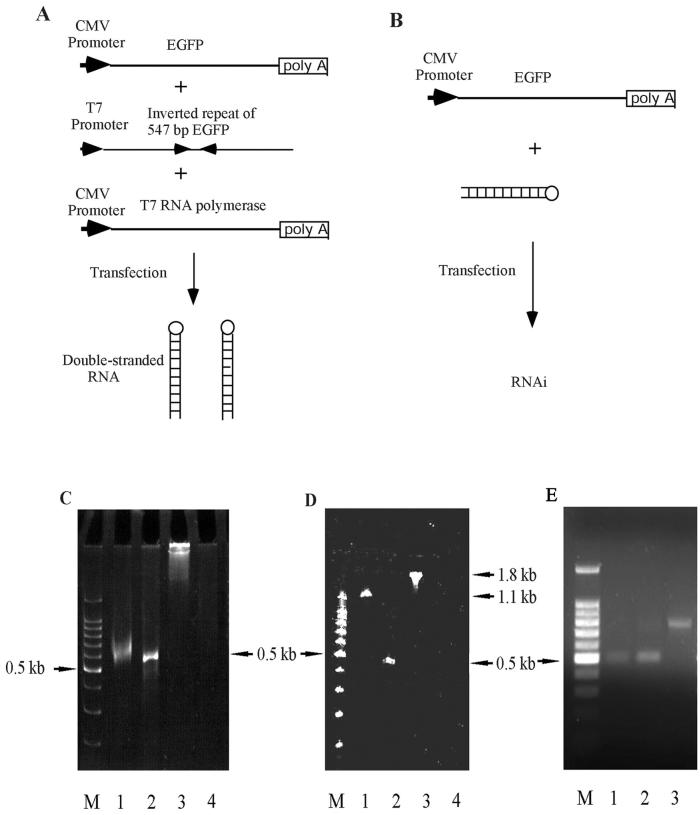
FIG. 1.
Strategy for generation of dsRNA. (A) In situ production of dsRNA of EGFP. An inverted repeat of EGFP, starting from the ATG codon to nucleotide 547 in the coding region, was cloned into the pGEMT vector under control of the T7 promoter (pGEMT-dsEGFP). RNAi activity in mammalian cells was tested by transient transfection of three plasmids, the target plasmid encoding the EGFP gene, the linearized pGEMT-dsEGFP, and plasmid encoding the T7 RNA polymerase cDNA. (B) Direct transfection of dsRNA made by in vitro transcription. Mammalian cells were transfected with dsRNA and the target plasmid encoding the EGFP gene to compare the RNAi activities between in situ production of dsRNA and in vitro-transcribed dsRNA. The dsRNA-EGFP was made by in vitro transcription using the T7 RNA polymerase and the linearized pGEMT-dsEGFP plasmid. (C) Analysis of dsRNA-EGFP transcribed by the T7 RNA polymerase. The pGEMT-dsEGFP plasmid was linearized by PstI, located at the 3′ end of the inverted repeat, to generate a runoff transcript by the T7 RNA polymerase. The transcribed RNA was digested with a mixture of RNases A and T1. The RNA was analyzed in a 5% nondenaturing acrylamide gel. Lane M, 100-bp dsDNA ladder. Lanes 1 and 2 depict in vitro-transcribed RNA with and without RNase treatment, respectively. Lanes 3 and 4 show a control 1.8-kb RNA provided in the RiboMax kit, with and without RNase treatment, respectively. (D) The same RNA samples were electrophoresed on a 5% denaturing acrylamide gel containing 40% formamide and 7 M urea. (E) Analysis of the dsRNA-lacZ transcribed by the T7 RNA polymerase. The sense and antisense RNAs were transcribed from the plasmid linearized by SalI and annealed to make dsRNA. Lanes 1 and 2 depict in vitro-transcribed antisense and sense RNAs, and lane 3 depicts annealed dsRNA in a 1% agarose gel.