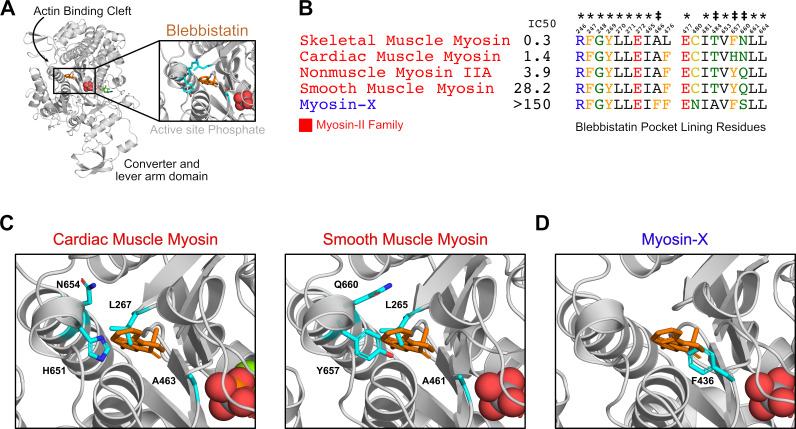
Figure 1. Blebbistatin’s cryptic binding pocket is closed in blebbistatin-free experimental myosin structures and the sequence of surrounding residues is highly conserved across myosin isoforms with widely varying IC50s.
(A) The motor domain of a D.d. myosin-II molecule bound to blebbistatin. The inset depicts blebbistatin’s cryptic binding pocket (PDB: 1YV324). Blebbistatin is shown in orange sticks while the active site phosphate is shown in spheres. Select residues (same as those shown in C) are shown in cyan sticks. (B) Alignment of blebbistatin contact residues (within 5 Å of blebbistatin in 1YV3) reveals that 16 of 19 residues are identical among myosin-IIs despite almost two orders of magnitude difference in blebbistatin IC50. We also include an unconventional myosin-X to highlight an important sequence difference at residue 466 (A vs. F) that separates blebbistatin-sensitive (IC50 <150 μM) and blebbistatin-insensitive isoforms (IC50 >150 μM). A star indicates a residue is conserved among all myosin isoforms shown. A double cross is used to indicate sequence differences previously suggested to play an important role in determining blebbistatin specificity (Allingham et al., 2005). (C) Crystal structures of β-cardiac (PDB: 5N6A Planelles-Herrero et al., 2017) and smooth muscle myosin (1BR2 Dominguez et al., 1998) do not suggest an obvious mechanism for differences in blebbistatin potency between these isoforms. In both structures, the cryptic blebbistatin-binding pocket is closed, and an aligned blebbistatin molecule (orange) clashes with a leucine residue (cyan). The two residues that differ between these isoforms (cyan histidine and asparagine in cardiac; cyan tyrosine and glutamine in smooth) do not form specific interactions with blebbistatin (e.g. hydrogen bonds). (D) In a blebbistatin-free myosin-X structure (PDB: 5I0H Ropars et al., 2016), F436 (cyan) forms a large steric impediment to blebbistatin binding (aligned molecule shown in orange) that is not present in myosin-II isoforms.