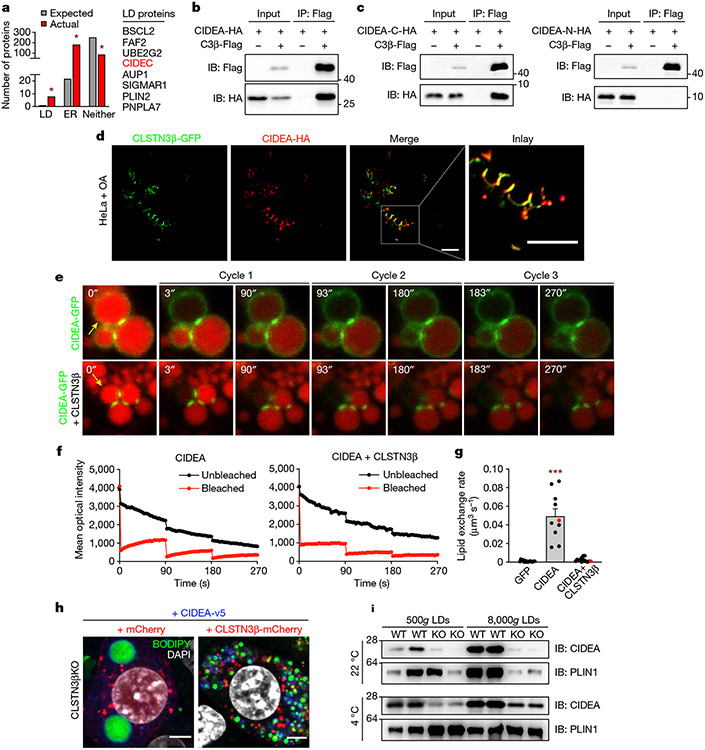
Fig. 5 ∣. CLSTN3β associates with CIDE proteins and blocks LD fusion.
a, Actual, number of proteins in the high-confidence CLSTN3β interactome with annotated LD, ER and neither localization in UniProt. Expected, number of proteins in a random sample of equal size from the UniProt mouse proteome with annotated LD, ER and neither localization. b, CLSTN3β-Flag and CIDEA-HA co-immunoprecipitation assay in transfected HEK293FT cells. c, CLSTN3β-Flag and HA-tagged CIDEA C-terminal region (CIDEA-C-HA) (left) or HA-tagged CIDEA N-terminal region (CIDEA-N-HA) (right) co-immunoprecipitation assay in transfected HEK293FT cells. d, Colocalization of CLSTN3β-GFP and CIDEA-HA in transfected HeLa cells treated with OA. Scale bar, 5 μm. e–g, FRAP-based lipid exchange rate assay in transfected 3T3-L1 preadipocytes incubated with 200 μM OA and 1 μg ml−1 BODIPY 558/568 C12 fatty acid for 16 h. e, Representative images of FRAP-based lipid exchange rate assay. Yellow arrows point to bleached LDs. f, Representative mean optical intensity traces for one pair of LDs from each group. g, Calculated lipid exchange rates (n = 15, 10, 14 cells). Cells used for representative traces are highlighted in red. h, Confocal microscopy of immortalized CLSTN3β KO brown adipocytes stably expressing mCherry or CLSTN3β-mCherry and CIDEA-v5 (day 5 of differentiation). Cells were stained with BODIPY 488, DAPI and anti-v5, and mCherry signal was amplified with anti-mCherry Alexa Fluor 594. Scale bar, 5 μm. i, BAT LDs isolated from female WT and AdC3KO mice housed at 22 or 4 °C for 24 h (n = 2 mice per lane). Equal amounts of TAG from each condition were loaded onto the gels. From the same experiment as Extended Data Fig. 6a,b. Bar plots show mean ± s.e.m. Each point represents a biological replicate. Two-sided *P < 0.001 by Fisher’s exact test (a), ***P < 0.001 by ordinary one-way ANOVA with Tukey’s multiple comparisons test (g).