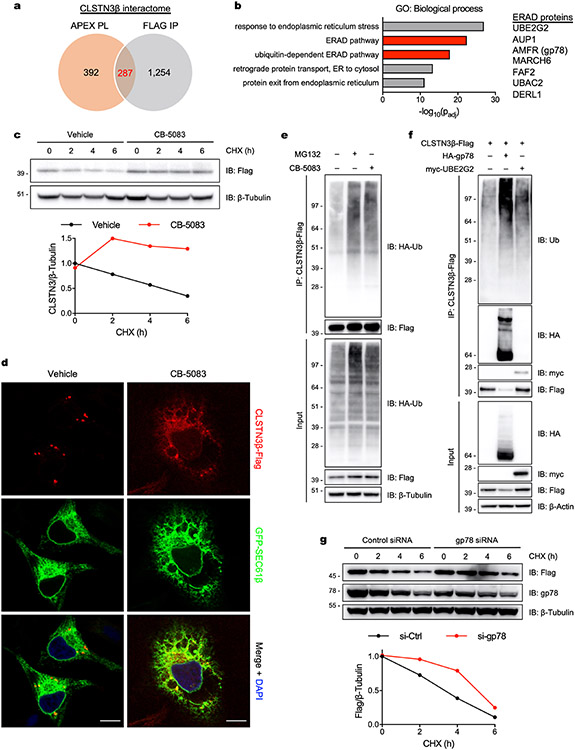
Extended Data Fig. 4 ∣. ER-localized CLSTN3β is degraded by ERAD.
a, Venn diagram showing the number of proteins identified as potential CLSTN3β binding partners by APEX proximity labeling (PL) and Flag immunoprecipitation (IP). b, (Left) Gene ontology (GO) analysis of the high-confidence CLSTN3β interactome (287 proteins). (Right) Partial list of ERAD proteins in the high-confidence CLSTN3β interactome. c, Immortalized brown adipocytes (day 6 of differentiation) were treated with cycloheximide (CHX, 100 μg/mL) for the indicated times and pretreated with either vehicle (DMSO) or CB-5083 (5 μM) for 30 min. d, Confocal microscopy of HeLa cells transfected with CLSTN3β-Flag and GFP-SEC61β and treated with either vehicle (DMSO) or CB-5083 (2.5 μM) for 8 h. Scale bar = 10 μm. e, CLSTN3β-Flag immunoprecipitation from HEK293T cells co-transfected with HA-ubiquitin and treated with either MG132 (25 μM) or CB-5083 (2.5 μM) for 4 h. f, CLSTN3β-Flag immunoprecipitation from HEK293T cells co-transfected with either HA-gp78 or myc-UBE2G2 and treated with CB-5083 (2.5 μM) for 4 h. g, HEK293T cells were co-transfected with CLSTN3β-Flag and either control or gp78 siRNA and treated with CHX (100 μg/mL) for the indicated times.