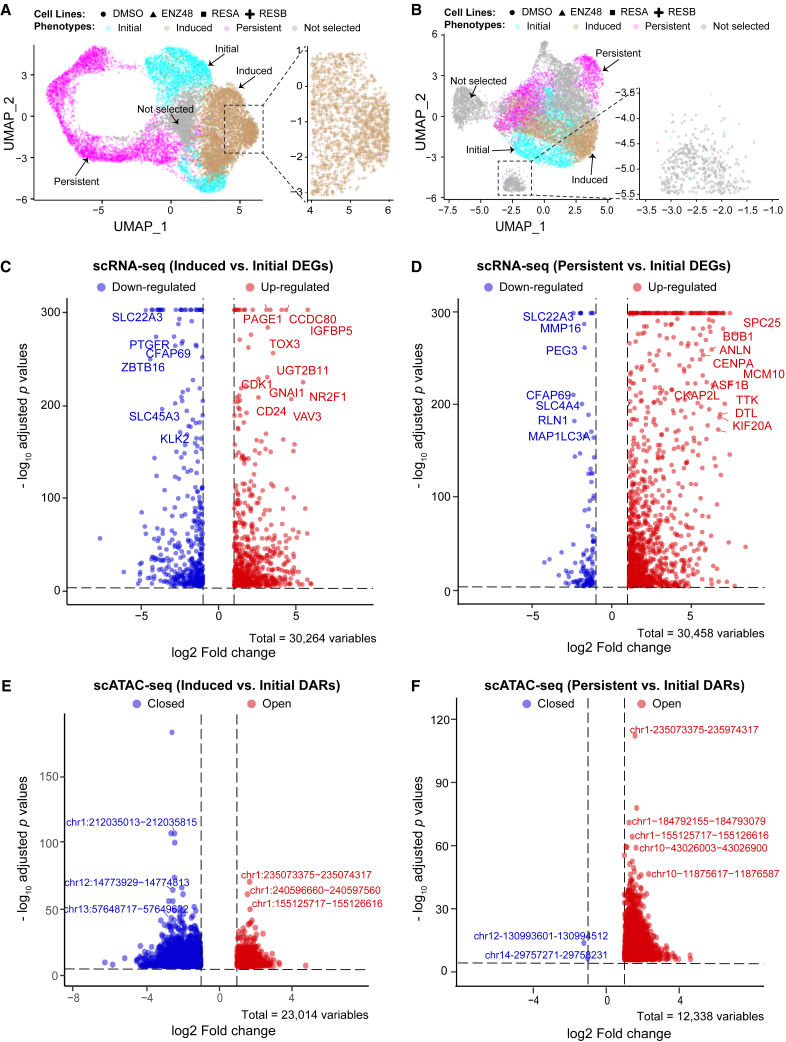
Figure 1.
ENZ-induced cellular phenotype shift
Dimension reduction uniform manifold approximation and projection (UMAP) plots are shown using scRNA-seq (A) and scATAC-seq data (B) in four PCa cell lines. Seurat clusters are colored according to their phenotypes as determined by their cellular compositions. Clusters with ambiguous classifications in gray are not selected for comparison analysis. Annotations for the color-coded phenotypes and shape-coded cell lines are on top of each plot. Volcano plots for comparisons between phenotypes of early response and initial and between that of persistent and initial are shown for scRNA-seq data (C and D) and for scATAC-seq data (E and F). Top differential events: differentially expressed genes (DEGs) or differentially accessible regions (DARs) are labeled on the plots.