Abstract
While considerable progress has been made in understanding the mechanisms of transcription in higher eukaryotes, transcription in single-celled, primitive eukaryotes remains poorly understood. Promoters of protein-encoding genes in the parasitic protist Trichomonas vaginalis, which represents one of the deepest-branching eukaryotic lineages, have a bipartite structure with gene-specific regulatory elements and a conserved core promoter encompassing the transcription start site. Core promoters in T. vaginalis appear to consist solely of a highly conserved initiator (Inr) element that is both a structural and a functional homologue of its metazoan counterpart. Using DNA affinity chromatography, we have isolated an Inr-binding protein from T. vaginalis. Cloning of the gene encoding the Inr binding protein identified a novel 39-kDa protein (IBP39). We show that IBP39 binds to both double and single Inr motifs found in T. vaginalis genes and that binding requires the conserved nucleotides necessary for Inr function in vivo. Analyses of the cloned IBP39 gene revealed no homology at the protein sequence level with identified proteins in other organisms or the presence of known DNA-binding domains. The relationship between IBP39 and Inr-binding proteins in metazoa presents interesting evolutionary questions.
The promoters of protein-encoding genes in metazoans have a bipartite structure, consisting of (i) a core promoter that encompasses the transcription start site and is directly responsible for the formation of the RNA polymerase II-containing preinitiation complex and (ii) binding sites for gene-specific regulators (34). Two common core promoter elements in higher eukaryotes are the TATA box, located 25 to 30 bp upstream of the transcription start site, and the initiator (Inr), a pyrimidine-rich element surrounding the start site. These elements are functionally analogous, since each is capable of directing accurate transcription initiation. In higher eukaryotes, core promoters have been shown to be quite heterogeneous; some contain both of these elements, while others contain either a TATA box or an Inr. In addition, for some genes the core promoter elements are undefined, as they lack either of these elements (reviewed in reference 28).
Despite significant progress in understanding the biochemical mechanisms of transcription in higher eukaryotes, the properties governing gene expression in deep-branching eukaryotic lineages remain poorly understood. The best studied of these organisms are the parasitic protists, for which recent studies have begun to define motifs critical for promoter function (8, 20, 22, 25, 29, 30, 33). The most striking result from these studies is the apparent lack of typical eukaryotic promoter elements, such as the TATA box or Inr. One group of parasites, the kinetoplastids, appears to lack sequence-specific promoters entirely. Instead, their protein-encoding genes are transcribed in large polycistronic units, and the process of trans-splicing regulates the formation of discrete mRNAs (reviewed in references 4 and 15). This lack of typical eukaryotic promoter elements may reflect the great divergence that has occurred since these organisms branched from the main line of eukaryotic descent.
Recently, we have examined the promoters of protein-encoding genes in the parasitic protist Trichomonas vaginalis (16, 17). This organism is a common human pathogen belonging to the phylum Parabasalia, one of the deepest-branching eukaryotic lineages (13). Trichomonads are characterized by a number of primitive cellular features, including the absence of two hallmark eukaryotic organelles, peroxisomes and mitochondria (5). Our studies have shown that promoters of protein-encoding genes in T. vaginalis, such as those of higher eukaryotes, consist of gene-specific regulatory elements and a conserved core promoter containing the transcription start site (16, 17). Core promoters in T. vaginalis appear to consist solely of a highly conserved Inr element whose consensus sequence, T C A + 1 Py (T/A), matches that of the metazoan Inr, Py Py A + 1 N (T/A) Py Py (17, 23, 27). This Inr has been found in all examined T. vaginalis genes and is both a structural and a functional homologue of the metazoan Inr. The T. vaginalis Inr is responsible for transcription start site selection, and its sequence requirements for activity are the same as those for metazoan Inr function (9, 17). The Inr, therefore, represents the first cognate core promoter element found in both deep-branching and metazoan eukaryotes, suggesting that it evolved early during eukaryotic evolution.
Since T. vaginalis appears to rely exclusively on the Inr to direct transcription initiation, it is likely that the trichomonad transcription machinery is highly optimized for Inr function. For this reason, we believe T. vaginalis is an excellent system in which to explore the mechanisms and evolution of Inr-mediated transcription in eukaryotes. A fundamental question in understanding Inr function in metazoans is the identity of the protein(s) that recognizes the Inr during transcription initiation. While several proteins, such as YY1, USF, TFII-I, and RNA polymerase II itself, have been shown to bind to sites that coincide with the Inrs of specific genes, TFIID has emerged as the prime candidate for recognizing the basal Inr during transcription initiation (reviewed in references 2, 3, and 26). In metazoans, the TFIID subunits TAFII150 and TAFII250 are required for Inr function and are in close contact with the core promoter DNA (11, 12, 21, 31, 32). In addition, a TAFII150-TAFII250 heterodimer has been shown to specifically recognize the Inr in a binding site selection assay, providing direct evidence of Inr binding by one or both of these TAFs (3). The exact mechanism of Inr binding, including the identity of the Inr-binding domain(s), remains to be defined.
To identify the protein(s) involved in Inr recognition in trichomonads, we have used DNA affinity chromatography to isolate an Inr-binding protein from T. vaginalis. This 39-kDa protein, IBP39, appears to be a novel DNA-binding protein, since database searches have failed to reveal homologues in other organisms. Using DNase I footprinting assays to assess the DNA-binding specificity of IBP39, we demonstrate that the sequence requirements for binding correspond to the conserved T. vaginalis Inr motif. Our results describe a previously unidentified, sequence-specific Inr-binding protein that is likely to be involved in promoter recognition by the T. vaginalis transcription machinery.
MATERIALS AND METHODS
Cell culture.
T. vaginalis strain C1 (ATCC 3001) was grown in Diamond's medium supplemented with 10% (vol/vol) horse serum and iron as described previously (23).
Plasmid construction.
The αSCS-CAT, Inr1, Inr2, Inr3, and Inr13 plasmids have been described previously (17). The FP plasmids (see Fig. 9) were generated by using the QuikChange site-directed mutagenesis method (Stratagene). Each mutagenesis reaction contained the αSCS-CAT plasmid as a template DNA and two complementary oligonucleotides, each containing the desired mutation surrounded by 15 bp of flanking sequence on both the 5′ and the 3′ sides. Constructs were verified by sequencing by using the Sequenase kit (USB).
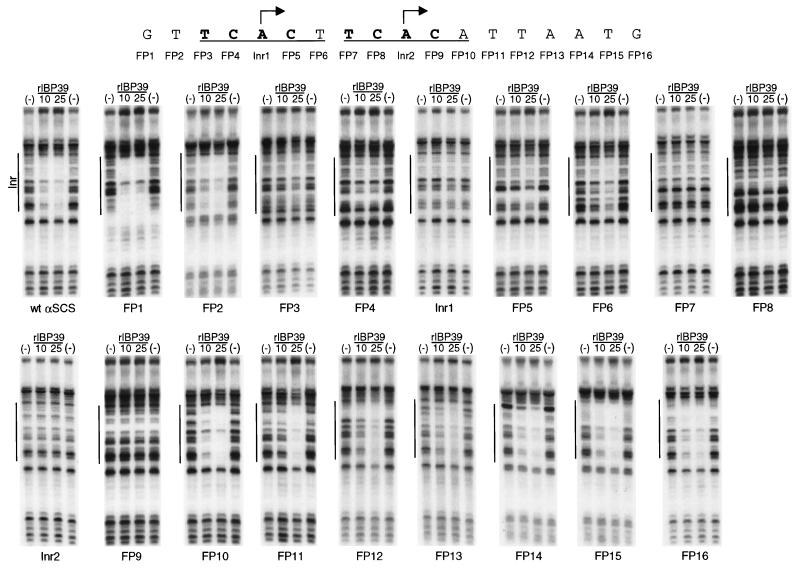
FIG. 9.
Recombinant IBP39 recognizes the initiator motif. DNase I footprinting assays were performed with recombinant IBP39 at either 10 or 25 ng and probes containing either the wild-type αSCS Inr or individual point mutations in the Inr region. The wild-type αSCS Inr region is shown. At each indicated position, a point mutation was introduced that altered both the type of base (pyrimidine versus purine) and the base pair (A-T versus G-C). The two Inr motifs on the probe are underlined, with the +1 positions indicated by arrows. Nucleotides which, when mutated, disrupted IBP39 binding are shown in boldface.
Preparation of the DNA affinity column.
Multimers of the wild-type αSCS Inr (Fig. 2A) were generated by a PCR-based method (7) by using the complementary primers wtInrA (5′-CTTGTTCACTTCACATTAATGCCCTTGTTCACTTCACATTAATGCC-3′) and wtInrB (5′-GGCATTAATGTGAAGTGAACAAGGGCATTAATGTGAAGTGAACAAG-3′). PCRs contained 50 ng of each primer, 1× PFU buffer [20 mM Tris-HCl (pH 8.8), 10 mM (NH4)2SO4, 2 mM MgSO4, 100 μg of bovine serum albumin/ml, 0.1% Triton X-100], a 10 μM concentration of each deoxynucleoside triphosphate, and Pfu polymerase. Cycling conditions were 95°C for 2 min, followed by 30 cycles of 95°C for 1 min, 65°C for 2 min, and 37°C for 1 min. PCR products were purified by phenol-chloroform extraction, followed by precipitation with ammonium acetate and ethanol. Each PCR yielded ∼6 μg of multimers that were between ∼200 and ∼5 kb in length. Multimer DNA (∼200 μg) was then bound to CNBr-activated Sepharose 4B (Pharmacia) according to the manufacturer's instructions.
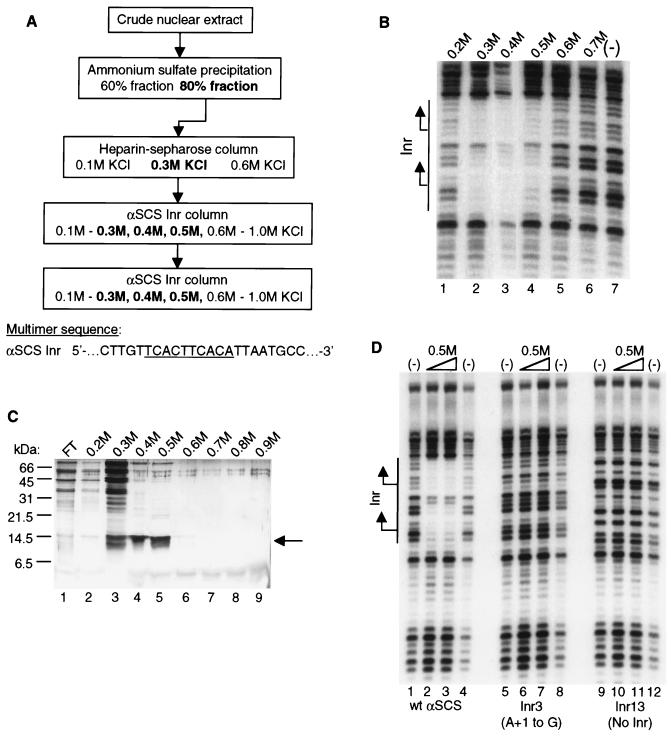
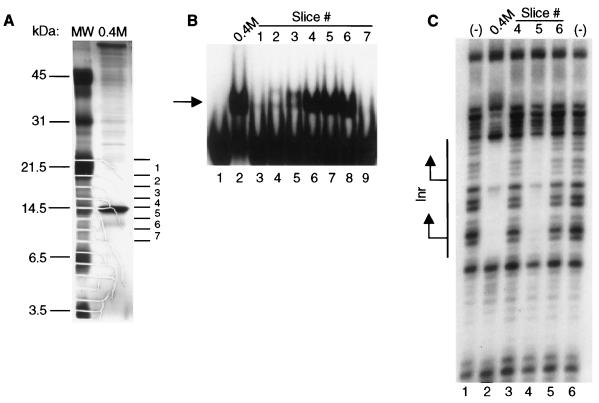
FIG. 2.
Purification of the initiator-specific binding activity. (A) Purification scheme for Inr-specific binding activity from T. vaginalis nuclear extracts. Column elutions were performed by using the indicated concentrations of KCl. The fractions that contained binding activity, as assessed by DNase I footprinting assays, are shown in boldface. The sequence of the αSCS Inr attached as a multimer to the DNA affinity columns is shown with the Inr motif underlined. (B) DNase I footprinting assay with fractions from the final αSCS Inr column. Footprinting reactions were performed with either the 0.2 to 0.7 M KCl αSCS Inr column fractions (lanes 1 to 6, respectively) or no protein (lane 7) and the wild-type αSCS Inr probe. No binding activity was detected in the flowthrough fraction (not shown). The DNA sequence of the Inr region of the probe in shown in Fig. 1. The location of the Inr element is shown with the two transcription start sites indicated by arrows. (C) Protein profile of fractions from the final αSCS Inr column. Protein from each fraction was precipitated with 25% TCA and separated on a 17.5% Tris-glycine SDS-PAGE gel, followed by silver staining. Lane 1, flowthrough fraction; lanes 2 to 9, 0.2 to 0.9 M KCl eluates from the αSCS Inr column. Positions of the molecular mass markers are indicated to the left. The 14.5-kDa protein band enriched in the 0.3 to 0.5 M KCl fractions is indicated by the arrow. (D) DNase I footprinting assay with the 0.5 M KCl eluate from the final αSCS Inr column. Footprinting reactions were performed with either no protein (lanes 1, 4, 5, 8, 9, and 12) or 5 μl (lanes 2, 6, and 10) or 10 μl (lanes 3, 7, and 11) of the 0.5 M KCl Inr column eluate and the wild-type αSCS Inr (lanes 1 to 4), Inr3 (lanes 5 to 8), or Inr13 (lanes 9 to 12) probes. The DNA sequence of the Inr region of the probes is shown in Fig. 1. The location of the Inr element is shown at the left with the two transcription start sites indicated by arrows.
Purification of IBP39.
Nuclear extracts from an 80-liter culture of T. vaginalis were prepared as described previously (17). The extracts were then fractionated by (NH4)2SO4 precipitation; (NH4)2SO4 was added to 60% saturation (0.36 g/ml), stirred for 45 min at 4°C, and then spun at 10,000 rpm in a JA-14 rotor (Beckman) for 45 min at 4°C. To the supernatant, additional (NH4)2SO4 was added to 80% saturation (0.13 g/ml) and precipitated as described above. The protein pellets were each resuspended in 40 ml of HGED0.1 (20 mM HEPES-KOH [pH 7.9], 100 mM KCl, 1 mM EDTA, 1 mM dithiothreitol [DTT], 10 μg of leupeptin/ml, 50 μg of TLCK [Nα-p-tosyl-l-lysine chloromethyl ketone]/ml, 1 mM phenylmethylsulfonyl fluoride, and 20% glycerol) and dialyzed against 4 liters of HGED0.1 for 8 h with three buffer changes. The 80% fraction contained 500 to 600 mg of total protein.
Next, the 80% (NH4)2SO4 fraction was applied to a 35-ml heparin-Sepharose column (Pharmacia) in HGED0.1. After a wash with 0.1 M KCl, 40 to 60 mg of total protein eluted with HGED0.3 (0.3 M KCl). The 0.3 M KCl heparin-Sepharose fraction was then dialyzed against 4 liters of HGED0.1 for 4 h with one buffer change. For DNA affinity chromatography (10), NP-40, to a final concentration of 0.01%, and 9 mg of poly(dG-dC) (Pharmacia) were added to the 0.3 M KCl heparin-Sepharose fraction. After 10 min on ice, the fraction was spun at 10,000 rpm in a JA-20 rotor (Beckman) for 10 min at 4°C. The supernatant was then applied to a 2-ml αSCS Inr affinity column. After a wash with HGED0.1 containing 0.01% NP-40 (HGEDN0.1), proteins were eluted with successive one column volume steps of HGEDN containing 0.2, 0.3, 0.4, 0.5, 0.6, 0.7, 0.8, 0.9, and 1.0 M KCl. The 0.3 to 0.6 M KCl fractions were pooled, diluted to 0.1 M KCl with HGEDN0 (no KCl), and applied to a 1-ml αSCS Inr affinity column. The column was washed, and the bound proteins were eluted as described above. Fractions were assayed for gel shift and DNase I footprinting activity. Proteins were visualized on a 17.5% Tris-glycine SDS-PAGE (sodium dodecyl sulfate-polyacrylamide gel electrophoresis) gel, followed by silver staining.
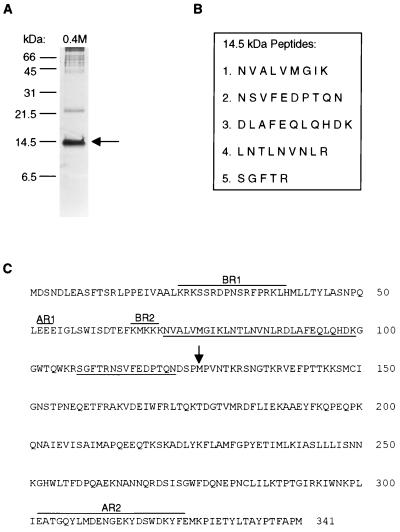
To isolate the 14.5-kDa protein for sequencing, the 0.4 M KCl second-pass Inr affinity column fractions from two purification runs were precipitated with 25% trichloroacetic acid (TCA) and separated on a 16.5% Tris-Tricine SDS-PAGE gel (24). Proteins were visualized by staining with Coomassie brilliant blue R 250, and the 14.5-kDa band was excised. The gel slices were washed twice with 50% acetonitrile and sent to the Harvard Microchemistry Facility (Cambridge, Mass.) for peptide sequencing. Approximately 5 μg of IBP39 was obtained in the 0.4 M KCl fractions from two purification runs.
Mobility shift assay.
Probes corresponding to −15 to +15 of the αSCS (Fig. 1) or ferredoxin Inrs (see Fig. 7) were prepared by annealing complementary oligonucleotides and end labeling them with [γ-32P]ATP and T4 polynucleotide kinase. The labeled probe was then purified on an 8% polyacrylamide gel. Unlabeled complementary primers containing the Inr3 and Inr13 mutations (Fig. 1) were annealed and used in competition assays. Each binding reaction contained the protein fraction, 5,000 cpm of labeled probe, 500 ng of poly(dG-dC), 20 mM HEPES-KOH (pH 7.9), 100 mM KCl, 1 mM EDTA, 1 mM DTT, 0.01% NP-40, and 10% glycerol. Reaction mixtures were incubated for 20 min at room temperature and run on a 6% 0.5× TBE polyacrylamide gel at 100 V for 2 h. For the supershift experiments, the binding reaction mixtures were preincubated with either preimmune or anti-IBP39 immunoglobulin G (IgG) for 30 min on ice prior to addition of the probe.
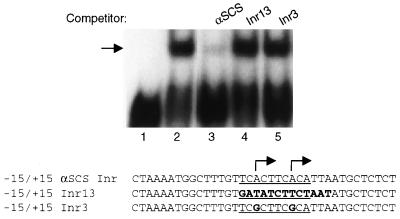
FIG. 1.
Initiator-specific binding activity in T. vaginalis nuclear extracts. A 37-bp probe, corresponding to positions −15 to +15 of the αSCS Inr region, was used in mobility shift assays with 20 μg of T. vaginalis crude nuclear extract. The αSCS Inr is a tandem Inr (underlined) that is known to contain two transcription initiation sites (indicated by the arrows) (17). Competition assays were performed with a 100× molar ratio of unlabeled wild-type and mutant αSCS Inr probes. Lane 1, no extract; lane 2, with extract; lane 3, competition with wild-type αSCS Inr; lane 4, competition with Inr13; lane 5, competition with Inr3. DNA sequences of the three probes are shown with the mutations in the Inr13 and Inr3 probes in boldface.
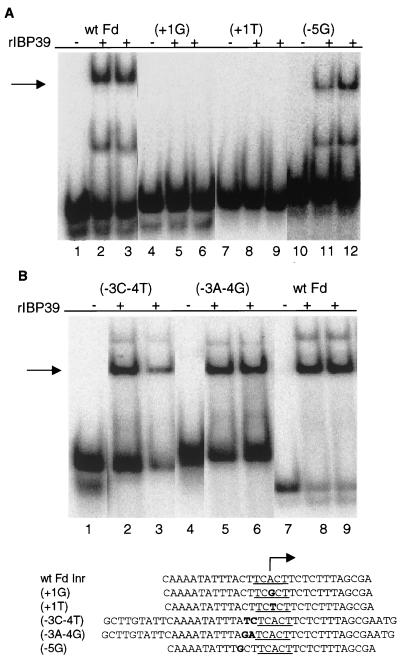
FIG. 7.
IBP39 recognizes a single functional Inr motif. Mobility shift assays were performed with 10 ng of recombinant IBP39 and wild-type and mutant ferredoxin Inr probes. The sequences of the probes are listed below. (A) Recombinant IBP39 binds the ferredoxin single Inr element. Lanes 1 to 3, wild-type ferredoxin Inr; lanes 4 to 6, +1 A-to-G mutation; lanes 7 to 9, +1 A-to-T mutation; lanes 10 to 12, −5 A-to-G mutation. Lanes 1, 4, 7, and 10 are DNA probe-only controls. (B) Mutation of non-Inr sequences just upstream of the ferredoxin Inr does not affect IBP39 binding. Lanes 1 to 3, −3 and −4 TC-to-CT mutation; lanes 4 to 6, −3 and −4 TC-to-AG mutation; lanes 7 to 9, wild-type ferredoxin Inr. Lanes 1, 4, and 7 are DNA probe-only controls.
DNase I footprinting assay.
The 184-bp probes were generated by PCR with the Δ7 primer (5′-ACTTACGCTTCAATTAAGGG-3′), which was end labeled with [γ-32P]ATP and T4 polynucleotide kinase; the CAT-PE primer (5′-TATATCAACGGTGGTATATCCAGTG-3′); and either the αSCS-CAT, Inr1, Inr2, Inr3, or Inr13 plasmids (17) or the FP1-16 plasmids (this study) as a template. Labeled probes were purified on a 6% polyacrylamide gel. DNase I footprinting reactions were then performed as described by Marshak et al. (18). Briefly, each 50-μl binding reaction mixture contained the protein fraction, 10,000 cpm of labeled probe, 10 mM HEPES-KOH (pH 7.9), 50 mM KCl, 1 mM EDTA, 1 mM DTT, 10% glycerol, and 0.2% polyvinyl alcohol. Reaction mixtures were incubated at room temperature for 20 min. Samples were then digested with either a 1:2,500 (for no-protein controls) or a 1:1,000 dilution of 2.5 mg of DNase I/ml, extracted with phenol-chloroform, ethanol precipitated, and separated on an 8% sequencing gel.
Denaturation-renaturation analysis.
Denaturation-renaturation analyses were performed as described by Baeuerle and Baltimore (1). Proteins were separated by SDS-PAGE, and slices were excised from the gel. Proteins from the gel slices were eluted, denatured, and renatured individually. Then, 10 μl of the renatured protein was used in each mobility shift reaction; 25 μl was used in each DNase I footprinting reaction.
Isolation of IBP39 genomic and cDNA clones.
To generate a fragment of the IBP39 gene, degenerate primers based on peptides 1 and 3 (Fig. 4B) were used in PCR with T. vaginalis genomic DNA. Use of the forward primer 5′-AACGTYGCYYTYGTYATGGG-3′ (corresponding to amino acids NVALVMG) and the reverse primer 5′-CTTRTCGTGYTGRAGYTGYTC-3′ (corresponding to amino acids EQLQHDK) resulted in a 90-bp product after PCR with 100 ng of T. vaginalis genomic DNA. PCR products were cloned into the pCR2.1-TOPO vector by using the TOPO-TA kit (Invitrogen) and sequenced with the Sequenase kit (USB). The PCR product was then labeled by random-primed synthesis with [α-32P]dATP and Klenow fragment and used to screen T. vaginalis cDNA and EcoRI genomic DNA libraries constructed in λZAPII (Stratagene). Plaques of positive clones were isolated, and pBluescript plasmids containing the genomic and cDNA clones were excised according to manufacturer's instructions. Clones with the largest genomic (1.6-kb EcoRI fragment) and cDNA (1,067-bp EcoRI-XhoI fragment) inserts were completely sequenced by the UCLA Sequencing/Genotyping Core Facility.
FIG. 4.
Amino acid sequence of IBP39. (A) Proteins from the 0.4 M KCl Inr column eluate were precipitated with 25% TCA and separated on a 16.5% Tris-Tricine SDS-PAGE gel, followed by silver staining. The arrow indicates the 14.5-kDa band excised for peptide sequence analysis. The positions of the molecular mass standards are shown on the left. (B) Peptide sequences obtained from tryptic fragments of the 14.5-kDa band. (C) Predicted amino acid sequence of IBP39. Amino acid residues matching those of the tryptic peptides derived from the purified IBP39 are underlined. The predicted proteolytic cleavage site at methionine 126 that resulted in the 14.5-kDa N-terminal peptide is marked by an arrow. Four regions enriched in either basic or acidic residues are overlined and indicated by BR1 and BR2 for the basic regions and AR1 and AR2 for the acidic regions.
Expression of recombinant IBP39.
To express recombinant IBP39, the coding region of the IBP39 gene was amplified from the cDNA clone by PCR by using the forward primer 5′-CACACCATGGATTCCAATGACCTTGAAGCAAGTTTTACATCTCGTC-3′, which contains an NcoI site, and the reverse primer 5′-CCCAGATCTCATTGGAGCGAAAGTAGG-3′, which contains a BglII site. The PCR products were cloned into the pCR2.1-TOPO vector (Invitrogen), and then the 1-kb NcoI-BglII fragment was subcloned into the pQE60 expression vector (Qiagen). IBP39 was then expressed as a C-terminal six-histidine fusion protein in Escherichia coli M15(pREP4) cells and purified by nickel column chromatography according to the manufacturer's instructions (Qiagen). To express the recombinant 14.5-kDa polypeptide, the coding region of IBP39 starting with amino acid 1 and ending with amino acid 126 was amplified by PCR by using the same forward primer as the full-length sequence and 5′-CCCAGATCTCATCGGGGAATCGTTTTG-3′ as the reverse primer. Expression and purification of the 14.5-kDa polypeptide were performed as described above.
IBP39 antisera and immunoblots.
Rabbit antiserum to IBP39 was prepared by Animal Pharm Services, Inc. (Healdsburg, Calif.), by using the recombinant IBP39-6 histidine fusion protein described above. For use in mobility shift assays, IgG from preimmune and anti-IBP39 antisera was purified by using protein A-Sepharose (Pharmacia) as recommended by the manufacturer. Nuclear extracts were prepared in the presence of protease inhibitors (Complete Mini Protease Inhibitor Cocktail [Roche], supplemented with 10 μg of leupeptin and 50 μg of TLCK/ml). Extracts were then separated by SDS–12% PAGE, followed by immunoblotting with anti-IBP39 antisera (used at a 1:4,000 dilution) and detection with protein A-horseradish peroxidase (1:3,500 dilution) and enhanced chemiluminescence reagents (Amersham).
Nucleotide sequence accession number.
The sequence of IBP39 has been deposited with GenBank under the accession number AF409099.
RESULTS
T. vaginalis nuclear extracts contain an Inr-specific binding activity.
As a first step toward the identification of T. vaginalis proteins that recognize the Inr, we identified an Inr-specific binding activity in T. vaginalis nuclear extracts (Fig. 1) (17). In an electrophoretic mobility shift assay with a 37-bp DNA probe containing the tandem Inr sequences (TCACTTCACA) from the α-succinyl coenzyme A synthetase (αSCS) gene (14), a strong binding activity was detected (Fig. 1, lane 2). This protein-DNA complex was abolished when an unlabeled competitor DNA containing a wild-type αSCS Inr was included but not when competitor DNAs lacking an Inr element (Inr13) or containing a specific mutation at the +1 positions (Inr3) was included (Fig. 1, lanes 3 to 5). Both of these Inr mutations have been shown previously to abolish Inr activity in vivo (17), indicating that this binding activity is specific for a functional Inr. No DNA-binding activity was observed by using either the coding or the noncoding strand of the 37-bp probe, demonstrating that the protein recognizing this motif binds only double-stranded DNA (data not shown). These data indicate that the binding activity observed in this assay is likely to play a role in Inr recognition in T. vaginalis.
Purification of the Inr-binding protein by DNA affinity chromatography.
We have purified the Inr-binding protein by using the strategy outlined in Fig. 2A. Briefly, T. vaginalis crude nuclear extracts were fractionated by ammonium sulfate precipitation, and the fraction containing the Inr-binding activity (80% ammonium sulfate) was subjected to heparin-Sepharose chromatography. The activity, which eluted in 0.3 M KCl, was then applied at a low salt concentration (0.1 M KCl) to a DNA affinity column containing multimers of a 23-bp sequence encompassing the αSCS Inr element. The αSCS Inr was used because it contains two Inr motifs in tandem (17), thus increasing the number of protein-binding sites on the column. After elution with higher KCl concentrations, fractions containing the binding activity were pooled, diluted to 0.1 M KCl, and applied to a second DNA affinity column. As shown in Fig. 2B, the Inr binding activity was detected by DNase I footprinting assays in fractions eluted from the second DNA affinity column with 0.3 to 0.5 M KCl. Strong protection of the DNA sequence surrounding the αSCS Inr was observed. This protection was centered over each of the two Inr motifs on the probe, and the binding was specific for a functional Inr element, since it was not seen with probes containing either a mutation of the +1 position (Inr3) or lacking an Inr sequence (Inr13) (Fig. 2D).
Examination of the eluted proteins by SDS-PAGE, followed by silver staining, revealed a prominent band at 14.5 kDa that was enriched in the 0.3 to 0.5 M KCl fractions (Fig. 2C). Several other prominent bands were also observed, especially in the 0.3 M fraction. However, none of these proteins was detected in all of the fractions that contained Inr-binding activity. To determine which of the protein bands was responsible for the Inr-binding activity, the 0.4 M KCl fraction was separated by SDS-PAGE and individual 4-mm gel slices spanning the gel lane were subjected to a denaturation-renaturation protocol (1) (Fig. 3A). The proteins recovered from each gel slice were tested for Inr-binding activity by mobility shift and DNase I footprinting assays. Protein recovered from gel slices 4 to 6, which contain and surround the 14.5-kDa band, were found to have Inr-binding activity in a mobility shift assay (Fig. 3B, lanes 6 to 8). Proteins recovered from gel slices of >21.5 kDa and <6.5 kDa did not have detectable Inr-binding activity (data not shown). In a DNase I footprinting assay, only protein recovered from gel slice 5 showed binding activity (Fig. 3C). Since this gel slice contains the bulk of the 14.5-kDa band and an equal volume of each eluate was added to the reactions, the lack of detectable binding recovered from gel slices 4 and 6 is likely to reflect the relatively low concentration of this protein in these slices. These results strongly indicated that the 14.5-kDa band was responsible for the Inr-binding activity. In addition, this band did not adhere to a DNA affinity column containing multimers of the +1 Inr mutation (data not shown), demonstrating that binding was specific to the wild-type Inr.
FIG. 3.
The 14.5-kDa protein band contains the Inr-binding activity. (A) Protein from the 0.4 M KCl final Inr column fraction was precipitated with 25% TCA and separated in duplicate lanes on a 17.5% Tris-glycine SDS-PAGE gel. Gel slices, ca. 4 mm in length, were excised from one lane for the denaturation-renaturation analysis, while the remainder of the gel was silver stained. The approximate position of each gel slice is indicated to the right of the silver-stained gel, and the molecular mass markers are shown on the left. (B) Gel shift assay with the −15/+15 αSCS Inr probe (Fig. 1) and either the 0.4 M KCl fraction (lane 2) or an equal volume of the eluate recovered from the individual gel slices following the denaturation-renaturation protocol (lanes 3 to 9). The Inr-specific binding activity is indicated by the arrow. (C) DNase I footprinting assay performed with either no protein (lanes 1, 6), the 0.4 M KCl fraction (lane 2), or an equal volume of the eluate recovered from gel slices 4, 5, and 6 (lanes 3 to 5, respectively) and the wild-type αSCS Inr probe. The location of the Inr element is shown with the two transcription start sites indicated by the arrows.
Cloning and characterization of the Inr-binding protein.
To identify the protein responsible for the Inr-binding activity, the 0.4 M KCl eluates from two purification runs were separated by Tris-Tricine SDS-PAGE to obtain increased resolution in the 14.5-kDa region (Fig. 4A) (24). The 14.5-kDa band was excised from the gel and subjected to proteolysis with trypsin. Five of the resulting peptides were then sequenced (Fig. 4B). To isolate the gene encoding the protein, degenerate primers based on the five peptide sequences were used in PCR with T. vaginalis genomic DNA. PCR with primers based on peptides 1 and 3 resulted in a 90-bp fragment that encoded peptides 1, 3, and 4. This PCR product was then used to screen a T. vaginalis genomic library and a clone containing a 1,024-bp open reading frame, encoding a 341-amino-acid, 39.3-kDa protein was obtained (Fig. 4C). This protein, which we have named IBP39 (for initiator binding protein, 39 kDa), has all five peptides obtained by microsequencing (underlined in Fig. 4C) clustered near the N terminus.
The size of IBP39 was unexpected as the Inr-binding protein was purified as a 14.5-kDa polypeptide. However, finding the exact same five peptides in the open reading frame of IBP39 as those derived from the 14.5-kDa protein makes it highly improbable that these sequences originate from different sources. Thus, the 14.5-kDa protein is either derived directly from the IBP39 gene or from a truncated duplication of this gene that retained the 5′ end and not the 3′ end. To distinguish between these possibilities, nonoverlapping DNA probes corresponding to amino acids 1 to 102 (N-terminal probe) or amino acids 210 to 294 (C-terminal probe) of IBP39 were hybridized with T. vaginalis genomic DNA. These data show that only one gene encodes both of these sequences (data not shown) and clearly establishes that the purified 14.5-kDa protein is a product of the IBP39 gene. In addition, Northern blot analysis of T. vaginalis mRNA using both the N-terminal and the C-terminal probes detected only a single 1-kb transcript, as predicted if only one gene gives rise to both sequences (data not shown). To confirm the size of the mRNA transcribed from the IBP39 gene and the sequence of IBP39 derived from our genomic clone, a T. vaginalis cDNA library was also screened with the 90-bp PCR product. This screen resulted in a cDNA clone containing the same open reading frame as the genomic clone, except that it lacked 21 bp at the 5′ end. Primer extension analysis with an IBP39 specific primer detected four transcription start sites from 3 to 10 bp upstream of the first ATG codon of the IBP39 open reading frame (data not shown). Since the most proximal ATG to the 5′ end of mRNAs is invariably used for translation initiation in T. vaginalis (A. Colocoussi and P. Johnson, unpublished data) and mRNA splicing has not been identified in this parasite, these results indicate that IBP39 is synthesized as a 341-amino-acid polypeptide.
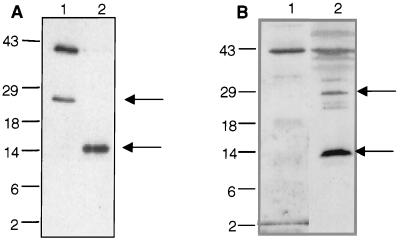
To examine the size of IBP39 in T. vaginalis, the recombinant protein was expressed in E. coli as a C-terminal six-histidine fusion protein, isolated by nickel column chromatography, and used to generate a polyclonal antiserum. Interestingly, a significant amount of recombinant IBP39 was cleaved during purification under nondenaturing conditions, as revealed by nickel agarose isolation of both the full-length 39-kDa protein and an abundant ∼25-kDa C-terminal peptide, both of which react with the anti-IBP39 antisera (Fig. 5A, lane 1). The N-terminal ∼14-kDa peptide, derived upon cleavage of IBP39, is not efficiently bound by the nickel column since it lacks the histidine tag. However, expression of a gene encoding only the first 126 amino acids of IBP39 (the region that contains the five peptide sequences derived from the 14.5-kDa protein) plus a histidine tag at the C terminus showed that this protein also has shared epitopes with IBP39 (Fig. 5A, lane 2). The detection of two breakdown products of recombinant IBP39, with molecular masses of ∼25 and ∼14.5 kDa, indicates that the cleavage of this protein occurs at a single site.
FIG. 5.
IBP39 exists as a 39-kDa polypeptide in vivo. (A) Anti-IBP39 antiserum recognizes full-length recombinant IBP39 and ∼25- and 14.5-kDa cleavage products from E. coli. Recombinant IBP39 was expressed as a C-terminal six-histidine fusion protein in E. coli and purified by nickel column chromatography in the absence of protease inhibitors. Nickel column eluates were then separated by SDS-PAGE, blotted, and reacted with the anti-IBP39 antiserum. Lane 1, nickel column eluate from cells expressing the full-length IBP39 six-histidine fusion protein; this protein and a degradation product of ∼25 kDa (marked by arrow) were purified. Lane 2, eluate from cells expressing the recombinant 14.5-kDa N-terminal peptide, corresponding to amino acids 1 to 126 of IBP39, with a C-terminal six-histidine tag. E. coli that expressed the 14.5-kDa peptide received the same treatment as those expressing full-length IBP39. (B) Western blot analysis of T. vaginalis nuclear extracts prepared in the presence of protease inhibitors. Lane 1, 25 μg of T. vaginalis freshly prepared nuclear extracts; lane 2, 25 μg of the same extract subjected to one freeze-thaw cycle at −80°C.
To determine whether the 14.5-kDa peptide purified from T. vaginalis is the result of posttranslational processing or proteolysis during extract preparation, we tested nuclear extracts prepared in the presence of a variety of protease inhibitor cocktails and found that a combination of calpains and serine, cysteine, and metalloprotease inhibitors resulted in the detection of almost exclusively the full-length 39-kDa protein (Fig. 5B, lane 1). However, a single freeze-thaw cycle of this extract at −80οC resulted in degradation of IBP39 to an abundant 14.5-kDa peptide (Fig. 5B, lane 2), while lesser amounts of the full-length IBP39 and the 25-kDa peptide were detected. These data confirm that the 14.5-kDa protein is a stable breakdown product of IBP39 resulting from degradation during purification. Moreover, these results strongly suggest that the endogenous T. vaginalis IBP39 exists primarily as a 39-kDa protein and does not undergo posttranslational processing in vivo.
IBP39 encodes a novel DNA-binding protein.
Extensive database searches using IBP39 failed to reveal significant homology to any known proteins in other organisms (for example, BLAST searches did not find any matches with E values of <0.6). Likewise, searches of protein domain databases revealed no previously identified protein domains. Thus, IBP39 appears to be a novel DNA-binding protein, based on its primary sequence and its lack of known DNA-binding domains. This protein is rich in leucine (8.5%) and lysine (9.1%) residues and has an overall basic charge, with a predicted pI of 8.4 (Fig. 4C). Secondary structure predictions using the GOR4 algorithm, indicated a mostly α-helical (42.5%) and random coil (44%) conformation for IBP39 (6). Two regions of IBP39 are predicted to be mostly α-helical, from amino acids 1 to 100 and 160 to 250, separated by a 60-amino-acid random coil region. This suggests that the structure of IBP39 may consist of two compact domains separated by an unstructured linker region. It is noteworthy that protease cleavage within this proposed linker region could result in an ∼14.5-kDa N-terminal domain, the form of IBP39 purified from T. vaginalis extracts (see arrow, Fig. 4A). IBP39 contains four regions enriched in charged amino acids (Fig. 4C). There are two basic regions near the N terminus, a 17-residue region from amino acids 23 to 39, and a cluster of four lysine residues from amino acids 66 to 70. There are also two acidic regions, a cluster of three glutamic acid residues at amino acids 52 to 54, and a 28-residue region from amino acids 302 to 329. Since the two basic regions are located within the N-terminal 14.5 kDa of IBP39, which likely contains the DNA-binding domain (discussed in detail below), they may be involved in DNA binding. The functional significance of the two acidic regions is unclear at present.
Recombinant IBP39 has Inr-specific binding activity.
To confirm that IBP39 has Inr-specific binding activity, the Inr-binding activity of recombinant IBP39 was tested in a DNase I footprinting assay (Fig. 6A). Recombinant IBP39 bound to the αSCS Inr element, resulting in a protection pattern similar to that seen with the 0.5 M Inr affinity column fraction (Fig. 6A, compare lanes 2 and 5). As with the purified protein, the binding of recombinant IBP39 was specific for a functional Inr; no binding was seen to a probe containing a mutation at the +1 position (Fig. 6A, lanes 8 to 11). This result shows that recombinant IBP39 has the same binding properties as the purified protein and is therefore most likely responsible for the Inr-binding activity in T. vaginalis extracts.
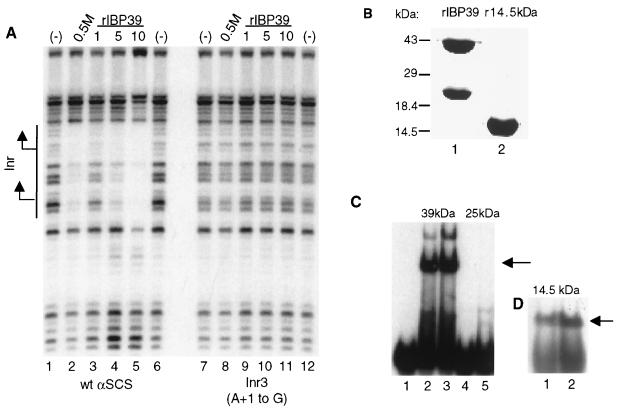
FIG. 6.
IBP39 specifically binds the initiator. (A) DNase I footprinting assays were performed with either the wild-type αSCS Inr (lanes 1 to 6) or the Inr3 (lanes 7 to 12) probes and either no protein (lanes 1, 6, 7, and 12), 5 μl of the 0.5 M KCl final Inr column fraction (lanes 2 and 8), 1 ng of recombinant IBP39 (lanes 3, 9), 5 ng of recombinant IBP39 (lanes 4 and 10), or 10 ng of recombinant IBP39 (lanes 5, 11). The location of the Inr element is shown with the two transcription start sites indicated by arrows. (B) Recombinant IBP39 (lane 1) and a 14.5-kDa N-terminal peptide corresponding to amino acids 1 to 125 of IBP39 (lane 2) were expressed in E. coli and separated on an SDS-PAGE gel. (C) Mobility shift assay using the −15/+15 αSCS Inr probe and the 39- and 25-kDa proteins recovered from the gel after the denaturation-renaturation protocol. Lane 1, no protein; lanes 2 and 3, either 1 or 5 μl, respectively, of the 39-kDa protein; lanes 4 and 5, either 1 or 5 μl, respectively, of the 25-kDa protein. (D) Mobility shift assay using the −15 or +15 αSCS Inr probe and 5 ng (lane 1) or 10 ng (lane 2) of the 14.5-kDa recombinant protein. The Inr-specific binding activities are indicated by the arrows.
As previously noted, a significant amount of recombinant IBP39 was cleaved during purification from E. coli. To confirm that the binding activity of IBP39 is confined to the N-terminal 14.5-kDa region, the full-length IBP39 and the C-terminal 25-kDa cleavage product were purified over a nickel column, separated on an SDS-PAGE gel, and used in a denaturation-renaturation assay (Fig. 6B). Proteins recovered from gel slices corresponding to IBP39 and the 25-kDa C-terminal peptide were then tested for Inr-binding activity in a mobility shift assay (Fig. 6C). In this assay, binding activity was detected with the full-length IBP39, but not with the 25-kDa C-terminal portion, indicating that the binding activity is contained exclusively within the first 14.5 kDa of IBP39. To directly test this, we expressed a 14.5-kDa recombinant protein containing only the first 125 amino acids of IBP39 (Fig. 6B, lane 2). In a gel shift assay, this 14.5-kDa protein also bound the Inr (Fig. 6D). These results are consistent with the purification of IBP39, using DNA affinity chromatography, as a 14.5-kDa polypeptide and confirm that the DNA-binding domain of IBP39 resides within the N-terminal 14.5 kDa of the protein.
Since IBP39 was purified by using the tandem Inr of the αSCS gene (Fig. 2A), we next tested whether this protein would also bind to the single Inr motif of the ferredoxin gene Inr (5′-CAAAATATTTACTTCACTTCTCTTTCGCGA-3′) in a mobility shift assay. As shown in Fig. 7A, recombinant IBP39 binds the wild-type ferredoxin Inr (lanes 1 to 3) but does not recognize this Inr when single mutations that disrupt its function (17) are introduced in the probe (lanes 4 to 9). The motif TTACT is found directly 5′ of the ferredoxin Inr. Although this motif resembles an Inr, there is no evidence that it functions as an Inr in vivo, since no ferredoxin mRNAs have 5′ ends mapping to this adenosine, using either primer extension or S1 nuclease assays (17, 23). Furthermore, this motif does not match the consensus motif of functional T. vaginalis Inrs (17). Nonetheless, to eliminate the possibility that this element might be necessary for IBP39 binding to the single ferredoxin Inr, we mutated the A (at −5 relative to the Inr) to a G. This mutation did not block IBP39 binding to the ferredoxin Inr, indicating that the single functional Inr motif is recognized (Fig. 7A, lanes 10 to 12). Additionally, we tested the effect on binding activity of mutating the nucleotides at −3 and −4 (C and T, respectively) in the ferredoxin Inr. Our previous work has shown that these positions are not needed for Inr activity in vivo (17). However, since these mutations disrupt the sequence CTTCAC, which is found in both the αSCS Inr and at nucleotides −4 to +2 in the ferredoxin Inr probe, this experiment was performed to eliminate the possibility that this motif was being recognized by IBP39 and to further confirm that the sequence immediately upstream of the ferredoxin Inr is not necessary for binding. Changing the nucleotides at −3 and −4 to either TC or AG had no or little effect on IBP39 binding (Fig. 7B, lanes 1 to 6) showing that IBP39 does not need this motif for binding. Collectively, these data show that IBP39 is capable of recognizing single Inr motifs and that binding is dependent on sequences known to be necessary for function. Determining whether IBP39 is capable of binding all single Inr motifs found in T. vaginalis genes will require further investigation.
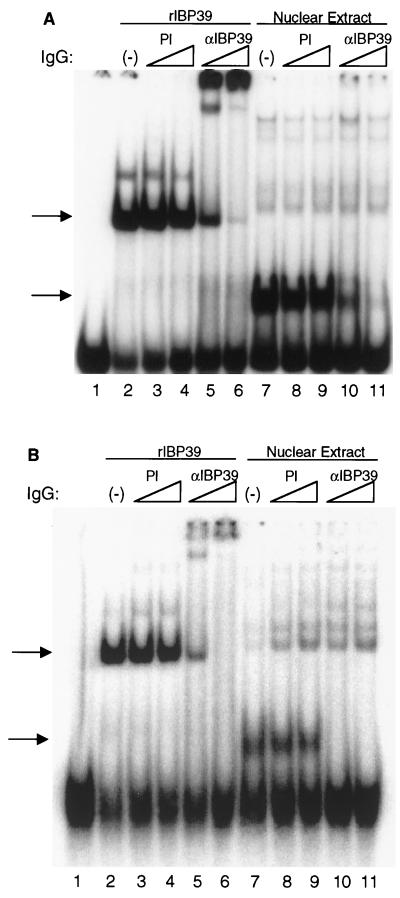
To directly test whether IBP39 was responsible for the Inr-binding activity seen in T. vaginalis extracts, IgG from preimmune and anti-IBP39 sera were purified by protein A-Sepharose chromatography and added to mobility shift reactions containing either recombinant IBP39 or T. vaginalis nuclear extracts and either the double αSCS Inr (Fig. 8A) or the single ferredoxin Inr (Fig. 8B). Addition of anti-IBP39 IgG to reactions containing either Inr probe and recombinant IBP39 resulted in a shift of the protein-DNA complex to the top of the gel (Fig. 8, lanes 5 and 6). When anti-IBP39 IgG was added to reactions containing T. vaginalis nuclear extracts, binding was abolished (Fig. 8, lanes 10 and 11). The addition of preimmune IgG had no effect on the bands detected with either the αSCS or ferredoxin Inr probes (Fig. 8, lanes 3 and 4 and lanes 8 and 9). These results clearly demonstrate that the protein in T. vaginalis nuclear extracts responsible for the Inr-specific binding activity was IBP39. The different effects of anti-IBP39 IgG in the mobility shift assay, i.e., supershift of the recombinant IBP39 complex versus inhibition of binding by the IBP39 in extracts, may result from the size difference between the two forms of IBP39. In the nuclear extract used for these experiments, IBP39 exists primarily as the 14.5-kDa N-terminal fragment containing the DNA-binding domain. Therefore, any epitopes recognized by the anti-IBP39 IgG are likely to block DNA binding. The recombinant IBP39, however, is full length and thus is likely to contain many epitopes which would not block DNA binding, resulting in a supershift of the protein-DNA complex.
FIG. 8.
IBP39 is responsible for the initiator-binding activity in T. vaginalis extracts. Mobility shift assays were performed with the −15/+15 αSCS (A) or the −15/+15 ferredoxin initiator (B) probes. Lane 1, no protein; lanes 2 to 6, 10 ng of recombinant IBP39; lanes 7 to 11, 12.5 μg of T. vaginalis nuclear extracts. Binding reaction mixtures were preincubated without probe with either no IgG (lanes 2 and 7); 1.5 μg (lanes 3 and 8) or 3.0 μg (lanes 4 and 9) of preimmune IgG; or 1.5 μg (lanes 5 and 10) or 3.0 μg (lanes 6 and 11) of anti-IBP39 IgG. The Inr-specific binding activities are indicated by the arrows.
IBP39 recognizes the Inr motif.
Previously, we have shown that the binding of IBP39 depends on the presence of an Inr motif and is blocked by mutations at the +1, −1, and −2 positions (Fig. 1 and 2D) (17). In order to define precisely the nucleotides recognized by IBP39, a series of individual point mutations were generated in the region containing the αSCS Inr. At each position, a mutation was introduced to alter both the type of base (pyrimidine versus purine) and the base pair (A-T versus G-C). This panel of mutations was then tested in DNase I footprinting assays with recombinant IBP39 to determine the effect of each mutation on DNA binding (Fig. 9). As expected, the mutations that disrupted IBP39 binding were located within both αSCS Inr motifs. In vivo analysis of T. vaginalis Inr function has shown that the most important nucleotides are a Py at −2, a C at −1, an A at +1, and a T or A at +3 (17). Likewise, mutation of these nucleotides disrupted recombinant IBP39 binding. Mutations at −2 (FP3 and -7), −1 (FP4 and -8) and +1 (Inr1 and Inr2) abolished binding by recombinant IBP39 (Fig. 9). Interestingly, mutations at the +3 position had less of an effect on IBP39 binding. While mutation of the 5′ +3 T to a G (FP6) greatly reduced IBP39 binding, mutation of the 3′ +3 A to a C (FP10) had little effect. This difference correlates with the different effects of these mutations on Inr activity in vivo: a G at the +3 position resulted in a reduction of Inr activity to 15% of wild-type, whereas a C at this position only reduced activity to 55% (17). The in vitro binding of IBP39 thus parallels in vivo Inr activity. Other positions in and around the Inr were also found to contribute to IBP39 binding. Mutation of the +2 position was found to abolish IBP39 binding in vitro (Fig. 9, FP5 and -9). This was somewhat surprising, since mutations at this position in vivo have a modest effect on Inr activity. For example, a +2 G mutation reduces Inr activity to 60% of that of the wild type (17). However, there does appear to be a strong preference for a pyrimidine at this position, since 28 of 33 Inr elements examined contained a T or C at this position (17). Pyrimidine nucleotides downstream of the Inr also appear to contribute to IBP39 binding. Mutation of the two T bases just downstream of the Inr motifs slightly reduced the binding of IBP39, as indicated by the weaker protection seen when 10 ng of protein was used (Fig. 9, FP11 and -12). This likely reflects a preference for pyrimidines surrounding the Inr, since 23 of 33 Inr elements examined have pyrimidines at both of these positions (17). Taken together, these results demonstrate that the sequence requirements for recombinant IBP39 binding are reflected in the nucleotide conservation of the Inr motif.
Individual point mutations disrupted IBP39 binding not only over the mutated Inr but also over the nonmutated motif, indicating that both tandem α-SCS Inr elements are required for binding in footprinting assays. This was unexpected since IBP39 binds the single ferredoxin Inr (Fig. 7) and most Inr elements in T. vaginalis contain only one Inr motif (17, 23). Moreover, our previous in vivo mutational analyses of αSCS Inr promoter activity showed that mutation of one of the two motifs reduced activity by only 60 to 80% (17). Additional experiments will be required to determine why binding to the αSCS promoter requires two Inrs in footprinting assays. One possible explanation is that the binding of two IBP39 proteins, one to each Inr motif, is necessary to stabilize DNA binding through protein-protein interactions, such that it is detectable in this assay. In vivo, however, IBP39 would be able to interact with other components of the T. vaginalis transcription machinery in order to stabilize its binding to the αSCS Inr.
DISCUSSION
This study describes the isolation and cloning of a novel Inr-binding protein, IBP39, from the protist T. vaginalis. IBP39 was isolated by DNA affinity chromatography as a 14.5-kDa polypeptide, derived from the N-terminal region of a 39-kDa protein. IBP39 was shown to have Inr-specific binding activity in both mobility shift and DNase I footprinting assays and is responsible for the binding activity in crude T. vaginalis nuclear extracts. Using a DNase I footprinting assay, we have also shown that the sequence requirements for IBP39 binding correspond to the conserved nucleotides of the T. vaginalis Inr element. Nucleotides that are critical for Inr function in vivo are also necessary for IBP39 binding. These results are consistent with a role for IBP39 in Inr recognition by the T. vaginalis transcription machinery.
Somewhat surprisingly, database searches with the IBP39 amino acid sequence have failed to identify homologues in other organisms. This suggests that IBP39 may represent a novel DNA-binding protein, one not yet identified in other eukaryotes. Since IBP39 is one of the first sequence-specific DNA-binding proteins isolated from a deep-branching eukaryote, it is not surprising that no closely related homologues have been reported from other protists. Furthermore, the great divergence between T. vaginalis and higher eukaryotes may obscure similarity at the protein sequence level to homologous sequence-specific DNA-binding proteins identified in higher eukaryotes. It is also possible that IBP39 homologues are not yet included in databases of eukaryotes whose genomic sequences are incomplete. Alternatively, a true functional homologue may not exist in other eukaryotes. We have also been unable to identify known DNA-binding domains within the IBP39 sequence. Again, this may reflect the great distance between T. vaginalis and higher eukaryotes, such that similarities at the sequence level are unclear, or IBP39 may contain a unique DNA-binding structure, one not yet identified in other eukaryotes.
Given the structural and functional similarities between their Inrs, trichomonads and metazoans may share a common mechanism for Inr recognition. In metazoans, TFIID is the main general transcription factor involved in core promoter recognition (reviewed in reference 26). The TFIID subunits TAFII150 and TAFII250 have been shown to be necessary for Inr-mediated transcription (12, 32), and a recent study has demonstrated direct Inr binding by a TAFII150-TAFII250 heterodimer (3). These results strongly suggest that these two TAFs are responsible for Inr recognition by TFIID. As noted above, similarity between IBP39 and either TAFII150 or TAFII250 is not apparent at the sequence level. However, since the DNA-binding domains of these two TAFs have yet to be identified, IBP39 may have limited homology to the Inr recognition domains of TAFII150 and/or TAFII250 that a computer search might miss. Alternatively, T. vaginalis may contain other, currently unidentified, Inr-binding proteins that may be homologues of various TFIID subunits. Another possibility is that IBP39 may represent a homologue of an as-yet-unidentified metazoan Inr-binding protein. Two studies have shown that TFIID is not sufficient to direct Inr-mediated transcription in a highly purified in vitro transcription system (12, 19). These studies identified several cofactors which were found to be required for Inr-mediated transcription: the CIF fraction, which contains the human homologue of TAFII150, among other proteins (12), and the TIC cofactors, which do not contain TAFII150 or the previously identified Inr-binding proteins YY1, USF, and TFII-I (19). Therefore, IBP39 may be homologous to a component of the CIF or TIC fraction which has yet to be characterized. Since all T. vaginalis core promoters appear to consist solely of an Inr, the trichomonad transcription machinery presumably does not have to distinguish between multiple core promoter elements, as does the metazoan transcription apparatus. Therefore, it is possible is that trichomonads have evolved a unique mechanism for Inr recognition, which is distinct from that of higher eukaryotes. Alternatively, IBP39 may be found to function like a TAF without sharing sequence homology with other eukaryotic TAFs.
The conservation of both structure and function of the Inr between trichomonads and metazoans strongly suggests that this core promoter element arose early during eukaryotic evolution. Interestingly, a sequence similar to the Inr is required for transcription of a gene in the protist Toxoplasma gondii, although it was not determined whether this element actually functions as an Inr (20). In addition, conserved sequences surround the transcription start sites of genes in the protists Entamoeba histolytica (22) and Giardia lamblia (30, 33). These elements do not share any sequence similarity with either each other or the trichomonad Inr. However, they may play a similar role during transcription initiation. This is the case for E. histolytica, since its Inr-like element has been shown to direct transcription start sites in vivo (25). Therefore, it is possible that these protists share a common mechanism for recognition and selection of transcription start sites with trichomonads. The identification of IBP39 represents the first step in characterizing the transcription machinery of T. vaginalis. Further work on the function of IBP39 and the identification of additional components of the trichomonad transcription apparatus should lead to a greater understanding of the evolution of gene regulation and the mechanisms of Inr function in eukaryotes.
ACKNOWLEDGMENTS
We thank members of the Johnson and Smale laboratories for helpful advice and discussion.
This work was supported by a grant from the NIH (AI30537) to P.J.J., a USPHS predoctoral training award (GM07185) to D.R.L., and a USPHS postdoctoral training award (AI07323) to A.O.T.L. S.T.S. is an Investigator with the Howard Hughes Medical Institute. P.J.J. is the recipient of a Burroughs-Wellcome Scholar Award in Molecular Parasitology.
REFERENCES
- 1.Baeuerle P A, Baltimore D. Activation of DNA-binding activity in an apparently cytoplasmic precursor of the NF-κB transcription factor. Cell. 1988;53:211–217. doi: 10.1016/0092-8674(88)90382-0. [DOI] [PubMed] [Google Scholar]
- 2.Burke T W, Willy P J, Kutach A K, Butler J E, Kadonaga J T. The DPE, a conserved downstream core promoter element that is functionally analogous to the TATA box. Cold Spring Harbor Symp Quant Biol. 1998;63:75–82. doi: 10.1101/sqb.1998.63.75. [DOI] [PubMed] [Google Scholar]
- 3.Chalkley G E, Verrijzer C P. DNA binding site selection by RNA polymerase II TAFs: a TAF(II)250-TAF(II)150 complex recognizes the initiator. EMBO J. 1999;18:4835–4845. doi: 10.1093/emboj/18.17.4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cross G A. Antigenic variation in trypanosomes: secrets surface slowly. Bioessays. 1996;18:283–291. doi: 10.1002/bies.950180406. [DOI] [PubMed] [Google Scholar]
- 5.Dyall S D, Johnson P J. Origins of hydrogenosomes and mitochondria: evolution and organelle biogenesis. Curr Opin Microbiol. 2000;3:404–411. doi: 10.1016/s1369-5274(00)00112-0. [DOI] [PubMed] [Google Scholar]
- 6.Garnier J, Gibrat J F, Robson B. GOR method for predicting protein secondary structure from amino acid sequence. Methods Enzymol. 1996;266:540–553. doi: 10.1016/s0076-6879(96)66034-0. [DOI] [PubMed] [Google Scholar]
- 7.Hemat F, McEntee K. A rapid and efficient PCR-based method for synthesizing high-molecular-weight multimers of oligonucleotides. Biochem Biophys Res Commun. 1994;205:475–481. doi: 10.1006/bbrc.1994.2690. [DOI] [PubMed] [Google Scholar]
- 8.Horrocks P, Dechering K, Lanzer M. Control of gene expression in Plasmodium falciparum. Mol Biochem Parasitol. 1998;95:171–181. doi: 10.1016/s0166-6851(98)00110-8. [DOI] [PubMed] [Google Scholar]
- 9.Javahery R, Khachi A, Lo K, Zenzie-Gregory B, Smale S T. DNA sequence requirements for transcriptional initiator activity in mammalian cells. Mol Cell Biol. 1994;14:116–127. doi: 10.1128/mcb.14.1.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kadonaga J T. Purification of sequence-specific binding proteins by DNA affinity chromatography. Methods Enzymol. 1991;208:10–23. doi: 10.1016/0076-6879(91)08004-2. [DOI] [PubMed] [Google Scholar]
- 11.Kaufmann J, Ahrens K, Koop R, Smale S T, Muller R. CIF150, a human cofactor for transcription factor IID-dependent initiator function. Mol Cell Biol. 1998;18:233–239. doi: 10.1128/mcb.18.1.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaufmann J, Verrijzer C P, Shao J, Smale S T. CIF, an essential cofactor for TFIID-dependent initiator function. Genes Dev. 1996;10:873–886. doi: 10.1101/gad.10.7.873. [DOI] [PubMed] [Google Scholar]
- 13.Keeling P J, Palmer J D. Parabasalian flagellates are ancient eukaryotes. Nature. 2000;405:635–637. doi: 10.1038/35015167. [DOI] [PubMed] [Google Scholar]
- 14.Lahti C J, Bradley P J, Johnson P J. Molecular characterization of the alpha-subunit of Trichomonas vaginalis hydrogenosomal succinyl CoA synthetase. Mol Biochem Parasitol. 1994;66:309–318. doi: 10.1016/0166-6851(94)90157-0. [DOI] [PubMed] [Google Scholar]
- 15.Lee M G, Van der Ploeg L H. Transcription of protein-coding genes in trypanosomes by RNA polymerase I. Annu Rev Microbiol. 1997;51:463–489. doi: 10.1146/annurev.micro.51.1.463. [DOI] [PubMed] [Google Scholar]
- 16.Liston D R, Carrero J C, Johnson P J. Upstream regulatory sequences required for expression of the Trichomonas vaginalis alpha-succinyl CoA synthetase gene. Mol Biochem Parasitol. 1999;104:323–329. doi: 10.1016/s0166-6851(99)00137-1. [DOI] [PubMed] [Google Scholar]
- 17.Liston D R, Johnson P J. Analysis of a ubiquitous promoter element in a primitive eukaryote: early evolution of the initiator element. Mol Cell Biol. 1999;19:2380–2388. doi: 10.1128/mcb.19.3.2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marshak D, Kadonaga J, Burgess R, Knuth M, Brennan W, Lin S. Strategies for protein purification and characterization. Plainview, N.Y: Cold Spring Harbor Press; 1996. pp. 127–303. [Google Scholar]
- 19.Martinez E, Ge H, Tao Y, Yuan C X, Palhan V, Roeder R G. Novel cofactors and TFIIA mediate functional core promoter selectivity by the human TAFII150-containing TFIID complex. Mol Cell Biol. 1998;18:6571–6583. doi: 10.1128/mcb.18.11.6571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakaar V, Bermudes D, Kyong R P, Joiner K A. Upstream elements required for expression of nucleoside triphosphate hydrolase genes of Toxoplasma gondii. Mol Biochem Parasitol. 1998;92:229–239. doi: 10.1016/s0166-6851(97)00220-x. [DOI] [PubMed] [Google Scholar]
- 21.Oelgeschlager T, Chiang C M, Roeder R G. Topology and reorganization of a human TFIID-promoter complex. Nature. 1996;382:735–738. doi: 10.1038/382735a0. [DOI] [PubMed] [Google Scholar]
- 22.Purdy J E, Pho L T, Mann B J, Petri W A., Jr Upstream regulatory elements controlling expression of the Entamoeba histolytica lectin. Mol Biochem Parasitol. 1996;78:91–103. doi: 10.1016/s0166-6851(96)02614-x. [DOI] [PubMed] [Google Scholar]
- 23.Quon D V, Delgadillo M G, Khachi A, Smale S T, Johnson P J. Similarity between a ubiquitous promoter element in an ancient eukaryote and mammalian initiator elements. Proc Natl Acad Sci USA. 1994;91:4579–4583. doi: 10.1073/pnas.91.10.4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schagger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 25.Singh U, Rogers J B, Mann B J, Petri W A., Jr Transcription initiation is controlled by three core promoter elements in the hgl5 gene of the protozoan parasite Entamoeba histolytica. Proc Natl Acad Sci USA. 1997;94:8812–8817. doi: 10.1073/pnas.94.16.8812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smale S T. Transcription initiation from TATA-less promoters within eukaryotic protein-coding genes. Biochim Biophys Acta. 1997;1351:73–88. doi: 10.1016/s0167-4781(96)00206-0. [DOI] [PubMed] [Google Scholar]
- 27.Smale S T, Baltimore D. The “initiator” as a transcription control element. Cell. 1989;57:103–113. doi: 10.1016/0092-8674(89)90176-1. [DOI] [PubMed] [Google Scholar]
- 28.Smale S T, Jain A, Kaufmann J, Emami K H, Lo K, Garraway I P. The initiator element: a paradigm for core promoter heterogeneity within metazoan protein-coding genes. Cold Spring Harbor Symp Quant Biol. 1998;63:21–31. doi: 10.1101/sqb.1998.63.21. [DOI] [PubMed] [Google Scholar]
- 29.Soldati D, Boothroyd J C. A selector of transcription initiation in the protozoan parasite Toxoplasma gondii. Mol Cell Biol. 1995;15:87–93. doi: 10.1128/mcb.15.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun C H, Tai J H. Identification and characterization of a ran gene promoter in the protozoan pathogen Giardia lamblia. J Biol Chem. 1999;274:19699–19706. doi: 10.1074/jbc.274.28.19699. [DOI] [PubMed] [Google Scholar]
- 31.Sypes M A, Gilmour D S. Protein/DNA crosslinking of a TFIID complex reveals novel interactions downstream of the transcription start. Nucleic Acids Res. 1994;22:807–814. doi: 10.1093/nar/22.5.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Verrijzer C P, Chen J L, Yokomori K, Tjian R. Binding of TAFs to core elements directs promoter selectivity by RNA polymerase II. Cell. 1995;81:1115–1125. doi: 10.1016/s0092-8674(05)80016-9. [DOI] [PubMed] [Google Scholar]
- 33.Yee J, Mowatt M R, Dennis P P, Nash T E. Transcriptional analysis of the glutamate dehydrogenase gene in the primitive eukaryote, Giardia lamblia. Identification of a primordial gene promoter. J Biol Chem. 2000;275:11432–11439. doi: 10.1074/jbc.275.15.11432. [DOI] [PubMed] [Google Scholar]
- 34.Zawel L, Reinberg D. Common themes in assembly and function of eukaryotic transcription complexes. Annu Rev Biochem. 1995;64:533–561. doi: 10.1146/annurev.bi.64.070195.002533. [DOI] [PubMed] [Google Scholar]