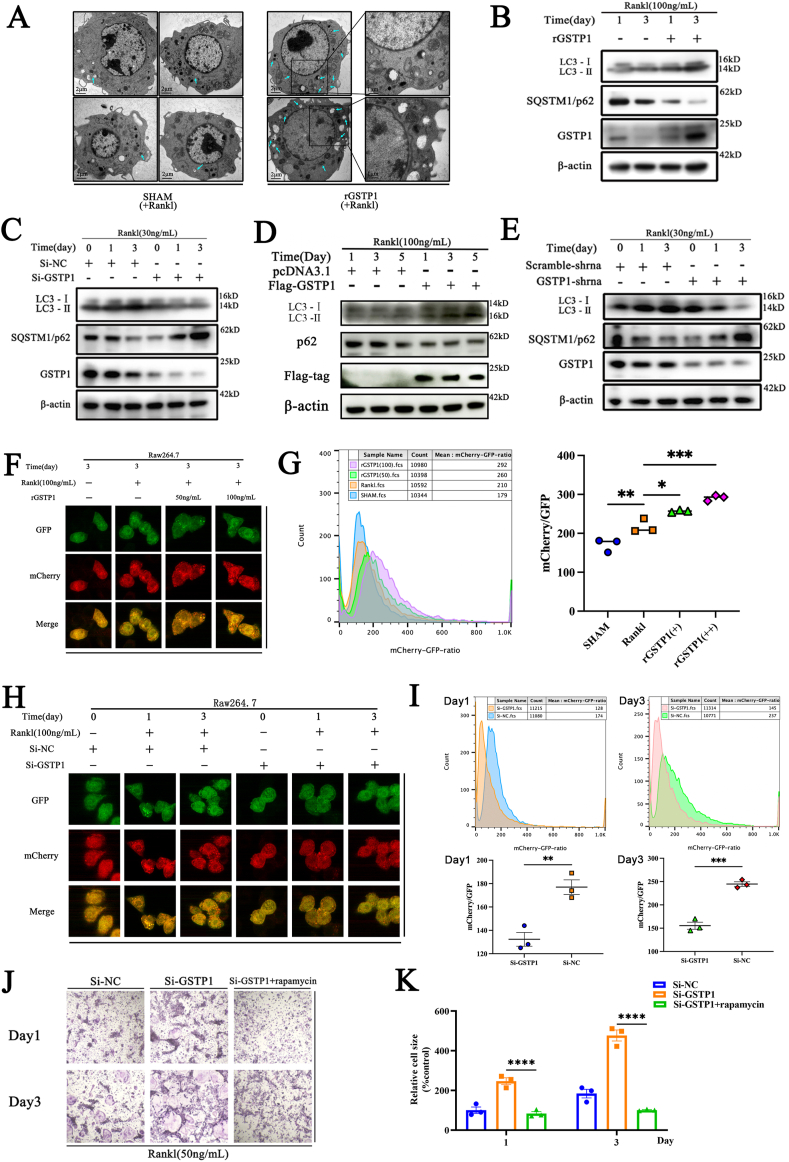
Fig. 3.
GSTP1 inhibits osteoclastogenesis through regulating autophagic flux. (A) TEM images of Raw264.7 cells differentiated with RANKL (100 ng/mL) stimulation in rGSTP1 group and SHAM group, green arrows represent autophagosomes and phagosomes. (B) WB analysis of the effects of rGSTP1 on LC-I, LC3-II and p62 protein expression during BMMs differentiation. (C) WB analysis of the effects of siRNA knockdown of GSTP1 on LC-I, LC3-II and p62 protein expression during BMMs differentiation. (D) WB analysis of the effects of overexpression of GSTP1 with Flag-GSTP1 plasmid on LC-I, LC3-II and p62 proteins expression during Raw264.7 differentiation. (E) Raw264.7 cells stably knocked down GSTP1 were treated with M-CSF and RANKL, WB analysis of LC-I, LC3-II and p62 proteins expression during Raw264.7 differentiation. (F) Raw264.7 cells stably transfected with mCherry-GFP-LC3 were constructed. Experiments were performed with different concentrations of rGSTP1, and cells were observed using a confocal microscope after stimulation with M-CSF and RANKL for 3 days. (G) Under the same conditions as (F), the ratio of mCherry/GFP in different groups was quantified by flow cytometry. (H) Raw264.7 cells stably transfected with mCherry-GFP-LC3 were constructed. After transfecting cells with Si-NC and Si-GSTP1, these cells were observed by confocal microscopy with M-CSF and RANKL for 3 days. (I) Under the same conditions as (H), the ratio of mCherry/GFP in different groups was quantified by flow cytometry. (J) The autophagy activator rapamycin reversed the staining profile and quantification of TRAP-positive cells promoted by low levels of GSTP1. (K) Quantitative analysis of cell size of nuclei of TRAP-positive cells.
All WB quantifications in this study were based on the grayscale values exhibited by the bands, all data are presented as mean ± SEM. Experimental data for each quantitative analysis were replicated at least three times. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)