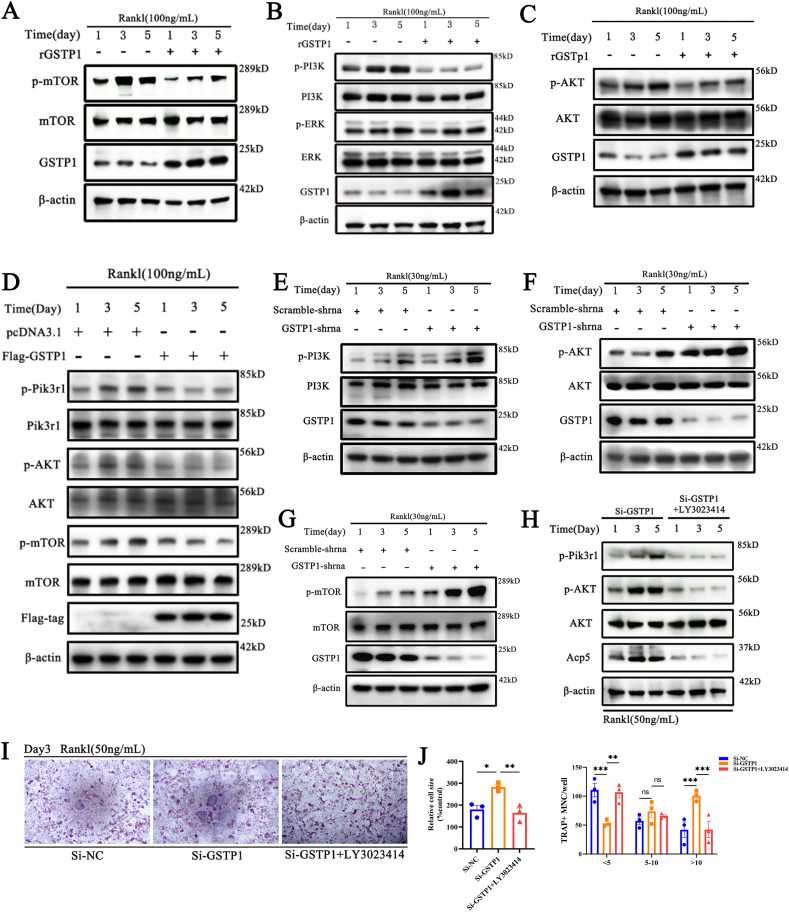
Fig. 4.
GSTP1 regulates autophagic flux through the Pik3r1/AKT/mTOR signaling axis. (A) BMMs were differentiated with M-CSF and RANKL (100 ng/mL) for 5 days, the WB results showed that rGSTP1(50 ng/mL) affects the expression of p-mTOR. (B) WB results showed that rGSTP1(50 ng/mL) affects the expression of p-Pik3r1, but not p-ERK1/2. (C) WB results showed that rGSTP1(50 ng/mL) affects the expression of p-AKT. (D) Raw264.7 cells were transfected with pcDNA3.1 or Flag-GSTP1 plasmid, and stimulated with M-CSF and RANKL (100 ng/mL) for 5 days. The WB results showed that the overexpression of GSTP1 regulated the level of p-Pik3r1, p-AKT, p-mTOR. (E) Raw264.7 cells transfected with shRNA-GSTP1 or shRNA-Scramble plasmids, and stimulated with M-CSF and RANKL (30 ng/mL) for 5 days. WB results show that low level of GSTP1 regulate the expression of p-Pik3r1. (F) WB results show that low level of GSTP1 regulate the expression of p-AKT. (G) WB results show that low level of GSTP1 regulate the expression of p-mTOR. (H) BMMs were differentiated with M-CSF and RANKL (50 ng/mL) for 5 days, One group used Si-GSTP1 and the other group used Si-GSTP1 + LY3023414 (10 nM, a non-toxic concentration, which alone did not inhibit osteoclast differentiation). WB results showed the differences in the expression of p-Pik3r1, p-AKT, AKT, ACP5 proteins between the two groups. (I) TRAP staining of Si-NC,Si-GSTP1 and Si-GSTP1+ LY3023414(10 nM) on the differentiation ability of BMMs under MCF and RANKL treatment. (J) Quantitative analysis of cell size and number of nuclei of TRAP-positive osteoclasts.
All WB quantifications in this study were based on the grayscale values exhibited by the bands, all data are presented as mean ± SEM. Experimental data for each quantitative analysis were replicated at least three times. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.