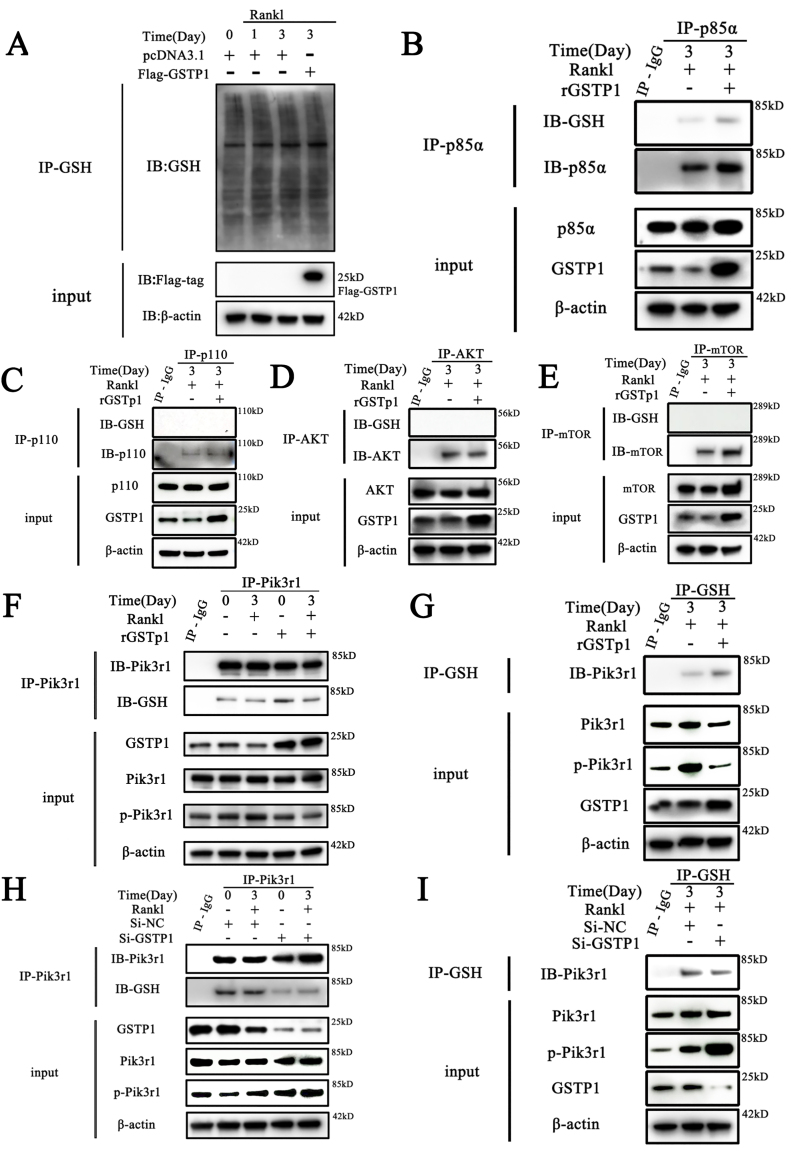
Fig. 5.
GSTP1 upregulates S-glutathionylation of Pik3r1 to inhibit its phosphorylation. The differentiation of BMMs was induced with M-CSF and RANKL, while rGSTP1 and Si-GSTP1 were used to up-regulate or down-regulate GSTP1 levels in cells, respectively. Then the related experiments are further completed. (A) S-glutathionylation levels of overall proteins on days 0, 1, 3 of osteoclast formation, and changes in S-glutathionylation levels when GSTP1 was upregulated. (B) Co-immunoprecipitation of Pik3r1, and WB analysis of S-glutathionylation levels. (C) Co-immunoprecipitation of p110, and WB analysis of S-glutathionylation levels. (D) Co-immunoprecipitation of AKT, and WB analysis of S-glutathionylation levels. (E) Co-immunoprecipitation of mTOR, and WB analysis of S-glutathionylation levels. (F) BMMs were treated with rGSTP1. Co-immunoprecipitation of Pik3r1, and WB analysis of Pik3r1 and GSH were used to detect the S-glutathionylation level of Pik3r1. (G) BMMs were treated with rGSTP1. Co-immunoprecipitation of GSH, and WB analysis of Pik3r1 was used to detect the S-glutathionylation level of Pik3r1. (H) BMMs were transfected with Si-GSTP1 and Si-NC. Co-immunoprecipitation of Pik3r1, and WB analysis of Pik3r1 and GSH were used to detect the S-glutathionylation level of Pik3r1. (I) BMMs were transfected with Si-GSTP1 and Si-NC. Co-immunoprecipitation of GSH, and WB analysis of Pik3r1 was used to detect the S-glutathionylation level of Pik3r1.
Co-IP findings are obtained from different blots using the same samples. All WB quantifications in this study were based on the grayscale values exhibited by the bands, all data are presented as mean ± SEM. Experimental data for each quantitative analysis were replicated at least three times. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.