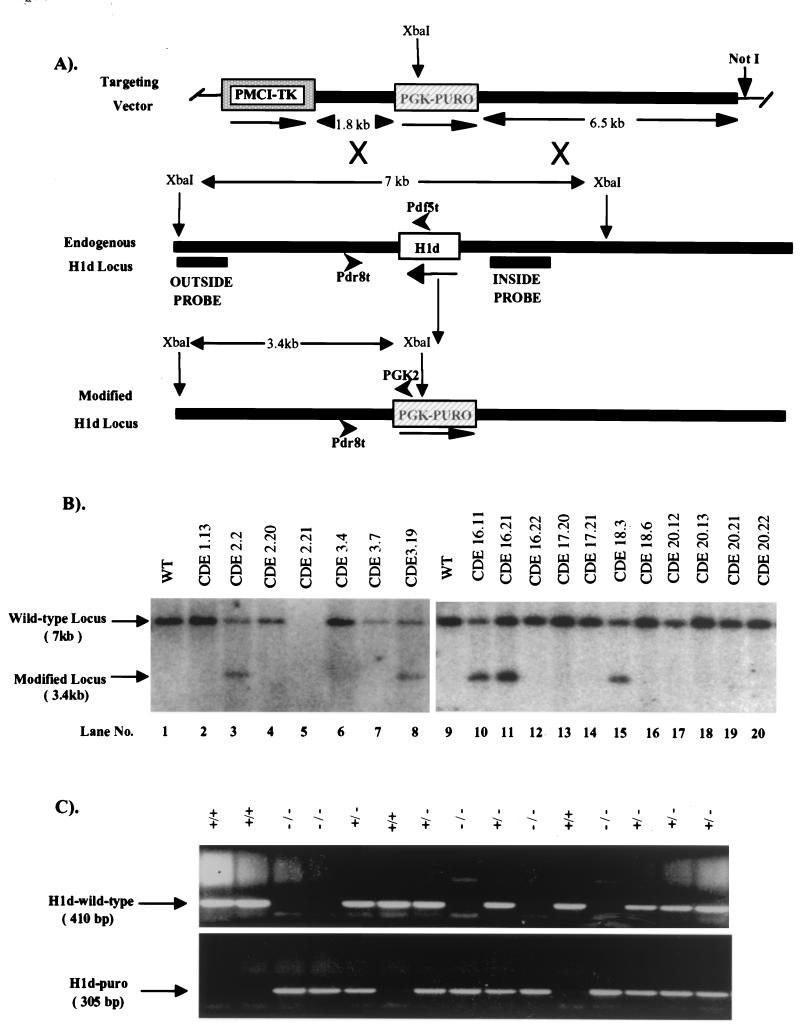
FIG. 2.
Targeted disruption of the H1d gene in mouse ES cells and mice. (A) Homologous recombination strategy in ES cells. To generate the H1d targeting vector (top), the Scramble plasmid system (Lexicon) was used. The positive and negative selectable marker genes PGK-PURO (striped box) and PMCI-TK cassettes (shaded box) were inserted into vector 901 at the AscI and RsrII sites, respectively. The H1d targeting vector (top) was then constructed by inserting a 6.5-kb H1d 5′ ClaI/EcoRI fragment (in which the EcoRI site lies 616 bp 5′ of the H1d ATG codon) and a 1.8-kb PCR-amplified H1d 3′ fragment (beginning 120 bp 3′ of the stop codon) into the SmaI/NotI and KpnI sites, respectively, in modified vector 901. A homologous recombination event (X's) between the targeting vector and the endogenous H1d locus (middle) results in production of a modified H1d locus (bottom), in which a 1.4-kb fragment, including the entire H1d coding sequence (open box), 616 bp of the 5′ noncoding sequence, and 120 bp of the 3′ noncoding sequence, is removed. (B) Identification of ES cell clones containing the modified H1d allele. ES cell DNA (10 μg) was digested with XbaI and blot hybridized with the outside probe (A). The expected positions of the hybridizing fragments from the unmodified (wild-type) and modified H1d loci and their respective sizes are indicated. Lanes 1 and 9 contained DNA from wild-type ES cells. Clones analyzed in lanes 3, 8, 10, 11, and 15 underwent a homologous recombination event. (C) Genotype analysis of offspring from parents heterozygous for the modified H1d allele. Siblings that were heterozygous for the modified H1d allele were bred, and 1 μg of tail DNA from offspring was used for PCR analysis with the following primers shown in panel A: H1d wild-type allele, the H1d 5′ sequence-specific primers (Pdf5t [5′ AAGCCTAAAGCTTCTAAGCCG 3′] and Pdr8t [5′ CTAGAGAACCCCCCTAATGC 3′]; predicted band size, 410 bp); H1d null allele (H1d-Puro): the PGK-PURO gene-specific primer (PGK2 [5′ GCTGCTAAAGCGCATGCTCCA 3′]) and the H1d 5′ sequence-specific primer (Pdr8t [5′ CTAGAGAACCCCCCTAATGC 3′]; predicted band size, 305 bp). The deduced genotype of each animal is indicated above each lane. The migration of PCR products from the wild-type and modified loci and their corresponding sizes are indicated.