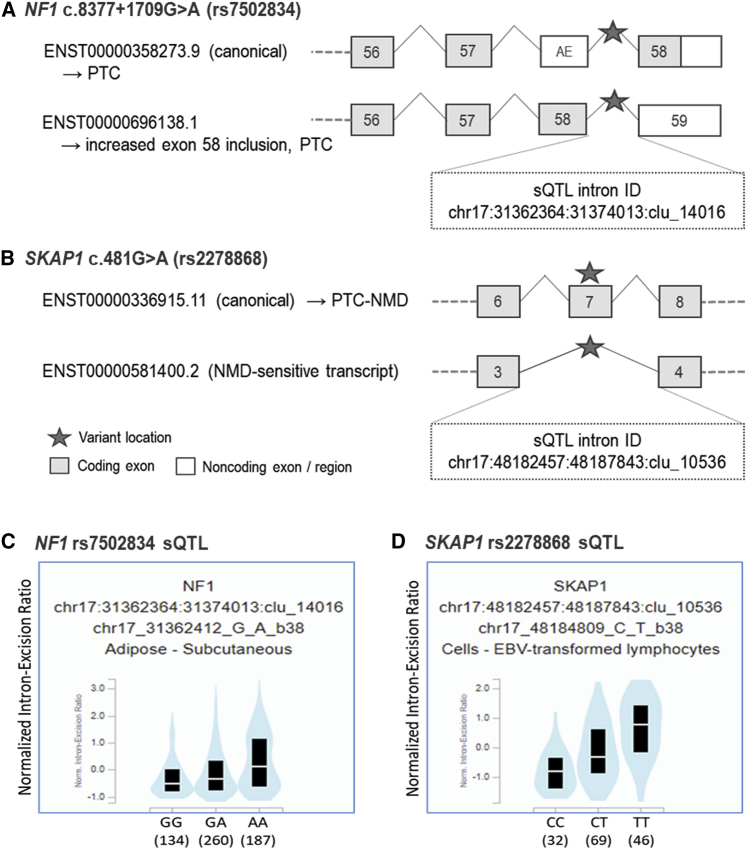
Figure 2.
rs7502834 (NF1) and rs2278868 (SKAP1) are predicted to affect splicing, and sQTL data demonstrate associations with corresponding splicing events
(A) and (B) show the predicted splicing events for rs7502834 and rs2278868 (denoted by the star symbols), respectively, with the corresponding intron IDs for the sQTLs. Vertically aligned exons have identical chromosomal locations, although the end of the 3′ untranslated regions may vary.
(C) and (D) show sQTL violin plots of normalized intron-exclusion ratios (GTEx v.8) for rs7502834 and rs2278868, respectively (see Table S3 for further details). Black boxes indicate interquartile ranges and the white lines show median values for each genotype. AE, alternative exon mapped to the canonical transcript; NMD, nonsense-mediated decay; PTC-NMD, premature terminating codon-nonsense-mediated decay; sQTL, splicing quantitative trait locus.