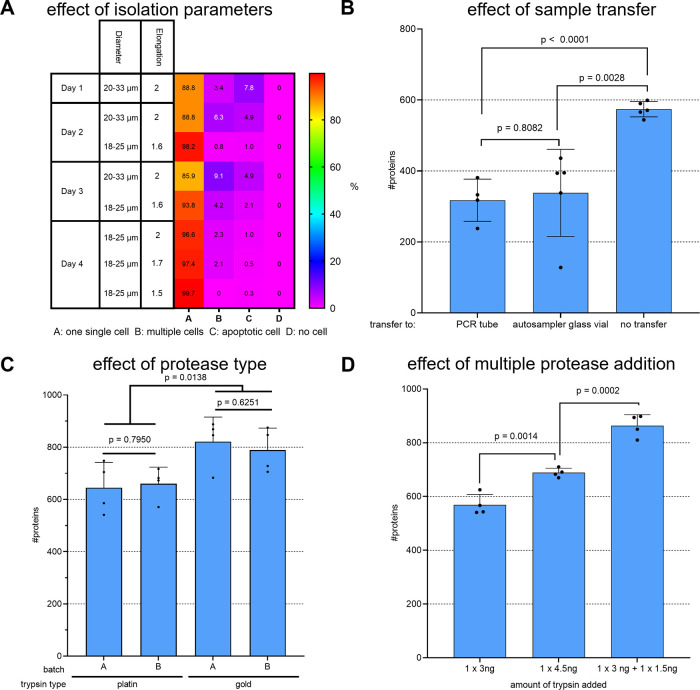
Figure 1.
Improving recovery and reproducibility by tuning workflow parameters. Individual HeLa cells were isolated into a predispensed master mix using CellenONE. (A) Parameters diameter and elongation were varied on CellenONE; numbers indicate the relative amount (%) of cells isolated in the category as indicated, n = 384 cells on 4 individual days each. (B) Individual HeLa cells were lysed and digested in a 384-well plate, and the sample was either injected from there to the LC-MS system or transferred as indicated (1 × 3 ng + 1 × 1.5 ng trypsin added, no DMSO was supplemented). (C) Cells were digested with two different batches of trypsin gold or platinum, each as indicated (1 × 3 ng + 1 × 1.5 ng trypsin added, no sample transfer, 5% DMSO supplemented). (D) Trypsin was added either once within the master mix (3 ng in 1 μL) or a second time by addition of 500 nL of additional fresh 3 ng/μL trypsin after 30 min. For comparison, the same total amount (4.5 ng in 1μL, middle bar) added in the beginning was included as the experimental condition (all conditions: without sample transfer and with 5% DMSO supplemented) (B–D) DDA data was analyzed using CHIMERYS at 1% FDR. The bars and error bars show the protein IDs obtained with their standard deviations. An unpaired two-tailed Student’s t-test was performed, and the resulting p-values are depicted above each compared data set, n ≥ 4 replicates.