Fig. 3. Structural characterization of ProteinMPNN designs.
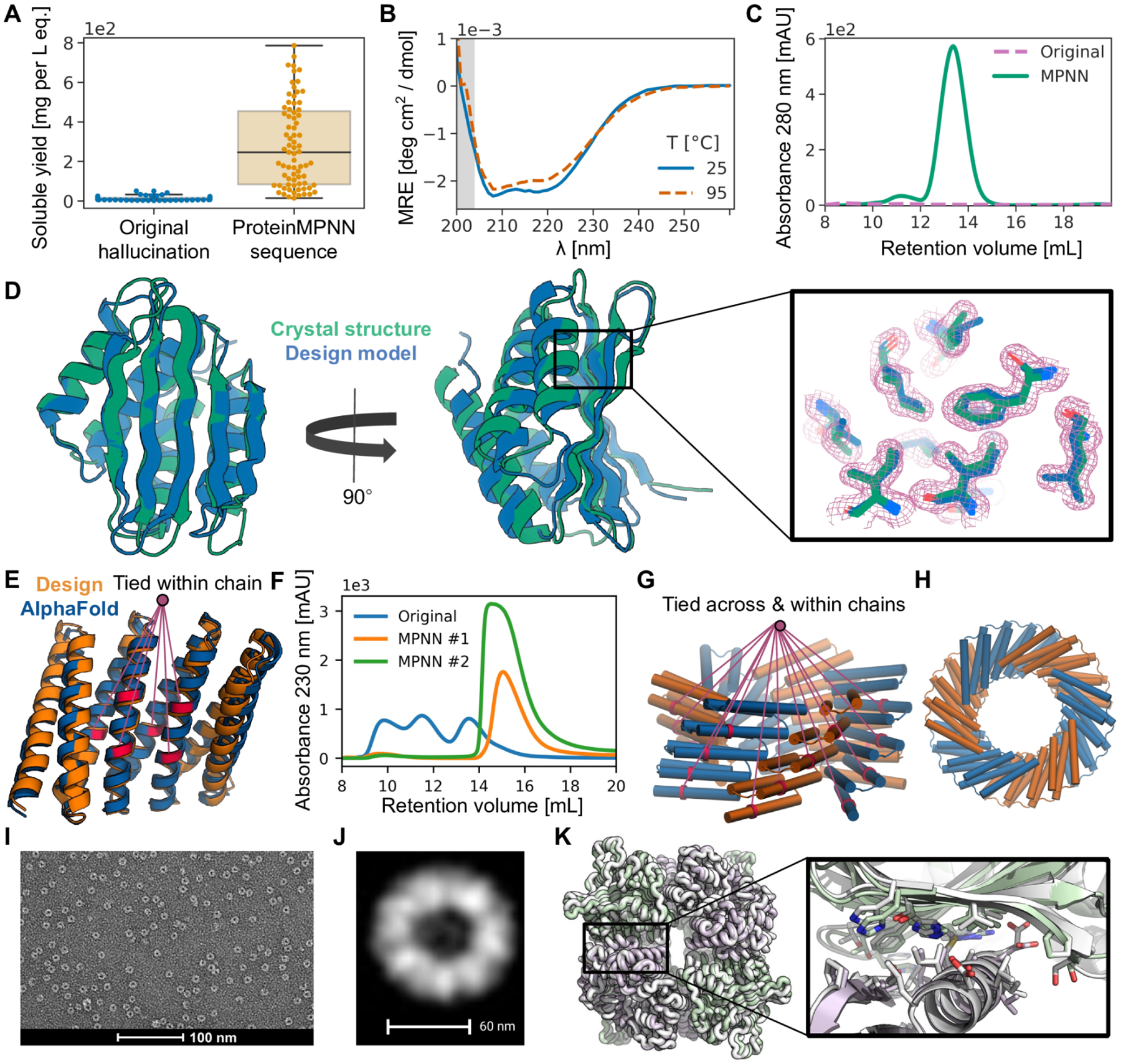
(A) Comparison of soluble protein expression over a set of AlphaFold hallucinated monomers and homo-oligomers (blue) and the same set of backbones with sequences designed using ProteinMPNN (orange), N=129. The total soluble protein yield following expression in E. coli, obtained from the integrated area unders size exclusion traces of nickel-NTA purified proteins, increases considerably from the barely soluble protein of the original sequences following ProteinMPNN rescue (median yields for 1 L of culture equivalent: 9 and 247 mg respectively). (B), (C), (D) In depth characterization of a monomer hallucination and corresponding ProteinMPNN rescue from the set in A. Like almost all of the designs in A, the sequence and structural similarity to the PDB of the design model are very low (E-value=2.8 against UniRef100 using HHblits, TM-score=0.56 against PDB). (B) The ProteinMPNN rescued design has high thermostability, with a virtually unchanged circular dichroism profile at 95 °C compared to 25 °C (C) Size exclusion (SEC) profile of failed original design overlaid with the ProteinMPNN sequence design, which has a clear monodisperse peak at the expected retention volume. (D) Crystal structure of the ProteinMPNN (8CYK) design is nearly identical to the design model (2.35 RMSD over 130 residues), see Figure S5 for additional information. Right panel shows model sidechains in the electron density, in green crystal side chains, in blue AlphaFold side chains. (E), (F) ProteinMPNN rescue of Rosetta design made from a perfectly repeating structural and sequence unit (DHR82). Residues at corresponding positions in the repeat unit were tied during ProteinMPNN sequence inference. (E) Backbone design model and MPNN redesigned sequence AlphaFold model with tied residues indicated by lines (~1.2Å error over 232 residues). (F) SEC profile of IMAC purified original Rosetta design and two ProteinMPNN redesigns. (G), (H) Tying residues during ProteinMPNN sequence inference both within and between chains to enforce both repeat protein and cyclic symmetries. (G) Side view of design model. A set of tied residues are shown in red. (H) Top-down view of design model. (I) Negative stain electron micrograph of purified design. (J) Class average of images from I closely match top down view in H. (K) Rescue of the failed two-component Rosetta tetrahedral nanoparticle design T33–27 (13) by ProteinMPNN interface design. Following ProteinMPNN rescue, the nanoparticle assembled readily with high yield, and the crystal structure (grey) is very nearly identical to the design model (green/purple) (backbone RMSD of 1.2 Å over two complete asymmetric units forming the ProteinMPNN rescued interface).
