Figure 1:
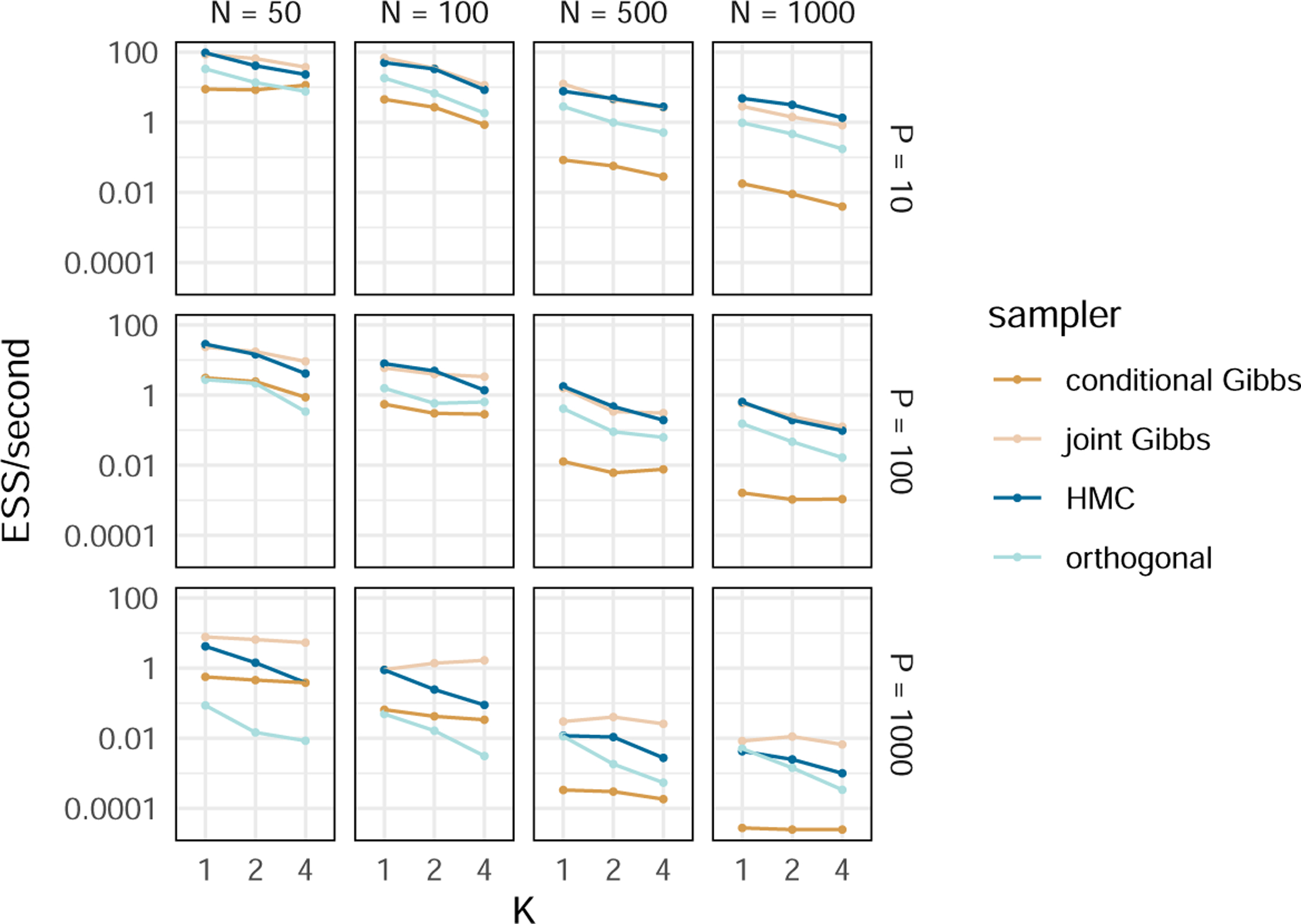
Timing comparison between inference regimes. We run three MCMC chain simulations for each combination of N (the number of taxa), P (the number of traits), K (the number of factors) and sampler and present the average minimum ESS per second for each. The “conditional Gibbs” sampler refers to the methods used by Tolkoff et al. (2017). The “joint Gibbs”, “HMC” and “orthogonal” samplers refer to the methods presented in Sections 3.1.1, 3.1.2 and 3.2 respectively. Our joint Gibbs and HMC samplers are an order of magnitude faster than the conditional Gibbs sampler with relatively few taxa (N = 50) but more than two orders of magnitude faster with many taxa (N = 1000). The orthogonal sampler is slower than the joint Gibbs and HMC samplers (and even the conditional Gibbs in the case of small-N, big-P) but scales well to large trees. Values are available in SI Table 1.
