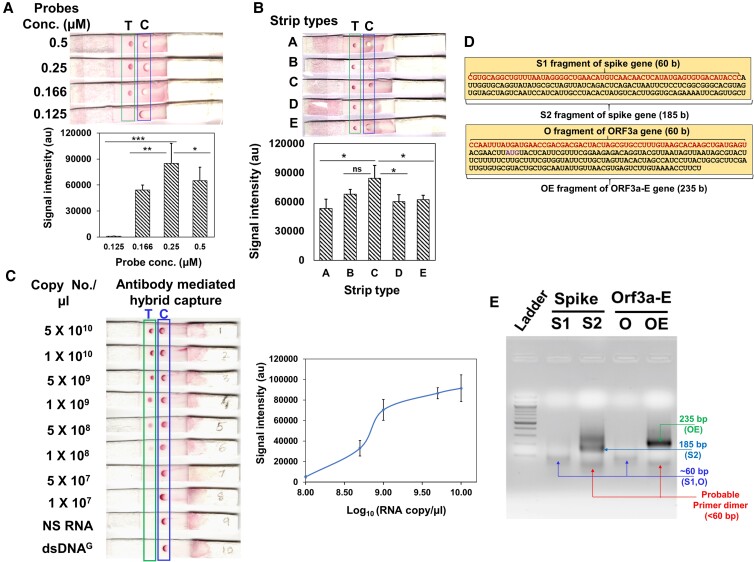
Fig. 3.
Proof of principle of RDH-LFA and TMIRA optimization. (A–C): Proof of principle of RDH-LFA:5×109 copies/µl of synthetic viral RNA with four biotinylated-DNA probes (12.5 nM) in 20 µl 1X hybridization buffer, unless otherwise indicated, were used for RDH-LFA; images showing results for different (A) probe conc. (B) strip type; the membrane wicking time in sec (membrane thickness in mm) are 150(110), 200(110), 100(105), 170(100), 120(105), for A, B, C, D, E type, respectively. (C) Image showing colorimetric signal developed in RDH-LFA for different conc. of the synthetic viral RNAs mixed with the four biotinylated-DNA probes (Table S1). NS RNA is non-specific RNA from human source. The signal intensity values from test spot are plotted against synthetic RNA conc. Also refer to Figure S2. (D) Target sequences to be amplified using TMIRA: different target lengths were tested for amplification efficiency [S1 (60 bp) and S2 (185 bp) for spike-gene, and O (61 bp) and OE (235 bp) for Orf3a-e gene]. (e) Agarose gel showing TMIRA products: lane 1 is 100 bp ladder, lane 2 and 3 are 60 bp (S1) and 185 bp (S2) amplicons using spike gene RNA template (S) with forward primer (T7-S-F) and reverse primers (S1-R for S1 and S2-R for S2, respectively), lane 4 and 5 are 61 bp (O) and 235 bp (OE) amplicons using Orf-3a-E gene RNA template with forward primer (T7-OE-F) and reverse primers (O-R for O and OE-R for OE, respectively). Different bands in the lanes are labeled with their respective sizes for better clarity.