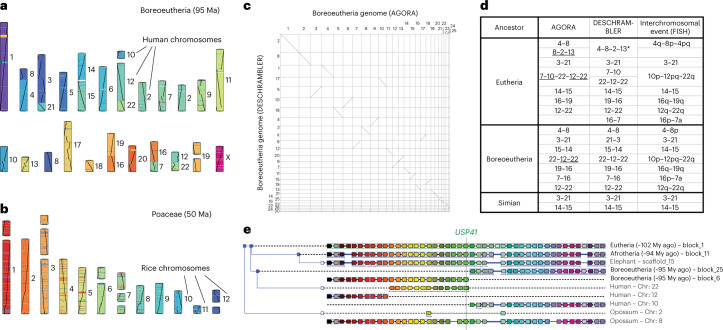
Fig. 4. AGORA ancestral genome reconstructions compared to extant genomes and state-of-the-art ancestral reconstructions.
a, The Boreoeutheria karyotype inferred by AGORA (the 25 largest CARs), coloured according to gene locations on human chromosomes, as indicated to the right of each CAR. b, The Poaceae karyotype inferred by AGORA (the 19 largest CARs), coloured according to gene locations on Oryza sativa chromosomes, as indicated to the right of each CAR. c, Collinearity of the Boreoeutheria ancestral genome reconstructed by AGORA with the genome reconstructed by DESCHRAMBLER33. d, Comparisons of computational reconstructions by AGORA and DESCHRAMBLER and Zoo-fluorescence in situ hybridization (FISH) linkage groups inferred for three key mammalian ancestors. Human chromosomes in ancestral linkage are indicated with hyphens. The Eutheria bolded linkage group 10–22–12 is documented in more detail in e. The underlined linkage groups are documented in Supplementary Fig. 14. DESCHRAMBLER reconstructed a linkage group between parts of human chromosomes 4, 8, 12 and 3 (asterisk) in disagreement with FISH evidence and AGORA when used on Ensembl v.92 data; however, this linkage group is also reconstructed by AGORA on Ensembl data v.102, suggesting an ambiguous ancestral linkage state (Supplementary Fig. 15). e, Gene adjacencies around the USP41 gene in extant species support the linkage of fragments of human chromosomes 10, 22 and 12 in the Eutheria ancestor. Orthologous genes are shown as arrows in matching colours, pointing in the direction of transcription. Opossum and elephant have both retained the ancestral organization at this locus, which has been rearranged in the human genome.