Figure 3.
ORF8/p62 condensates facilitate DMV formation
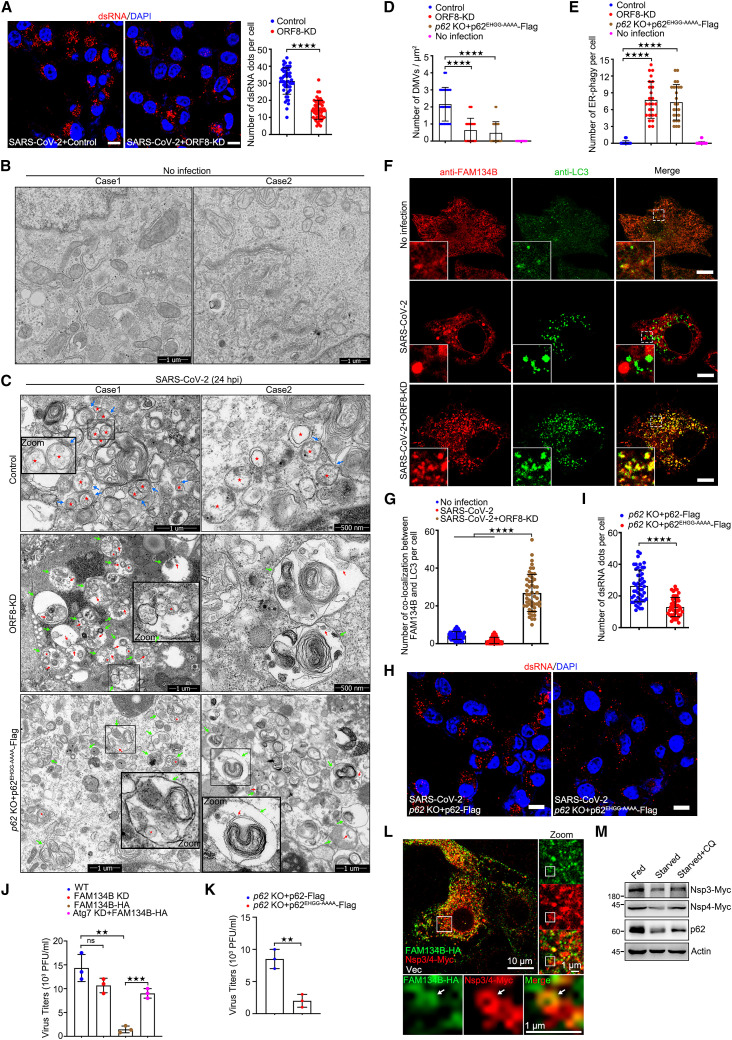
(A) Vero-E6 cells were transfected with control or ORF8-KD plasmid for 12 h and infected with SARS-CoV-2 for 24 h. Cells were stained with dsRNA antibody (red). Nuclear DNA was stained with DAPI (blue). Cells were imaged using confocal microscopy. Scale bar, 10 μm. The number of dsRNA puncta in each cell was counted from 50 cells of two independent experiments. Two-tailed unpaired Student’s t test, ∗∗∗∗p < 0.0001.
(B and C) Transmission electron microscopy images of p62 WT and KO Vero-E6 cells transfected with p62EHGG-AAAA with control or ORF8-KD plasmid for 12 h and then infected with or without SARS-CoV-2 for 24 h. The DMV structures (red pentagram), ER-DMVs interaction (blue arrowhead), autophagosomes (green arrowhead), ER in autophagosomes (red arrowhead) are shown.
(D and E) The number of DMV (D) and ER-phagy (E) in each cell was counted from 25 cells of two independent experiments. Two-tailed unpaired Student’s t test, ∗∗∗∗p < 0.0001.
(F) Vero-E6 cells were transfected with control or ORF8-KD plasmid for 12 h and then infected with or without SARS-CoV-2 for 24 h, then cells were stained with anti-FAM134B and anti-LC3 antibodies and imaged using confocal microscopy. Scale bar, 10 μm.
(G) The number of co-localizations between FAM134B and LC3 in each cell from (F) was counted from 25 cells of two independent experiments. Two-tailed unpaired Student’s t test, ∗∗∗∗p < 0.0001.
(H) p62 KO Vero-E6 cells transfected with WT or p62EHGG-AAAA for 12 h and then infected with SARS-CoV-2 for 24 h. Cells were stained with dsRNA antibody (red). Nuclear DNA was stained with DAPI (blue). Cells were imaged using confocal microscopy. Scale bar, 10 μm.
(I) The number of dsRNA puncta in each cell from (H) was counted from 50 cells of two independent experiments. Two-tailed unpaired Student’s t test, ∗∗∗∗p < 0.0001.
(J) Vero-E6 cells were transfected with sh-FAM134B or FAM134B-HA or si-Atg7 for 12 h, cells were infected with SARS-CoV-2 for 24 h, and media were collected and analyzed using plaque assay. Error bars, mean ± SD of three independent experiments. Two-tailed unpaired Student’s t test, ∗∗p < 0.01, ∗∗∗p < 0.001.
(K) p62 KO Vero-E6 cells were transfected with p62-FLAG or p62EHGG-AAAA-FLAG for 12 h and infected with SARS-CoV-2 for 24 h, and media were collected and analyzed using plaque assay. Error bars, mean ± SD of three independent experiments Two-tailed unpaired Student’s t test, ∗∗p < 0.01.
(L) HeLa cells were transfected with FAM134B-HA, Nsp3/4-Myc for 24 h, then stained with anti-HA and anti-Myc, and imaged using confocal microscopy. Scale bar, 10 μm.
(M) HeLa cells were transfected with Nsp3/4-Myc for 24 h and starved for another 12 h with or without 2 h CQ treatment. Cell lysates were analyzed using WB.
See also Figure S4.