Figure 2: Metabolic requirements for NRF2 sensitivity.
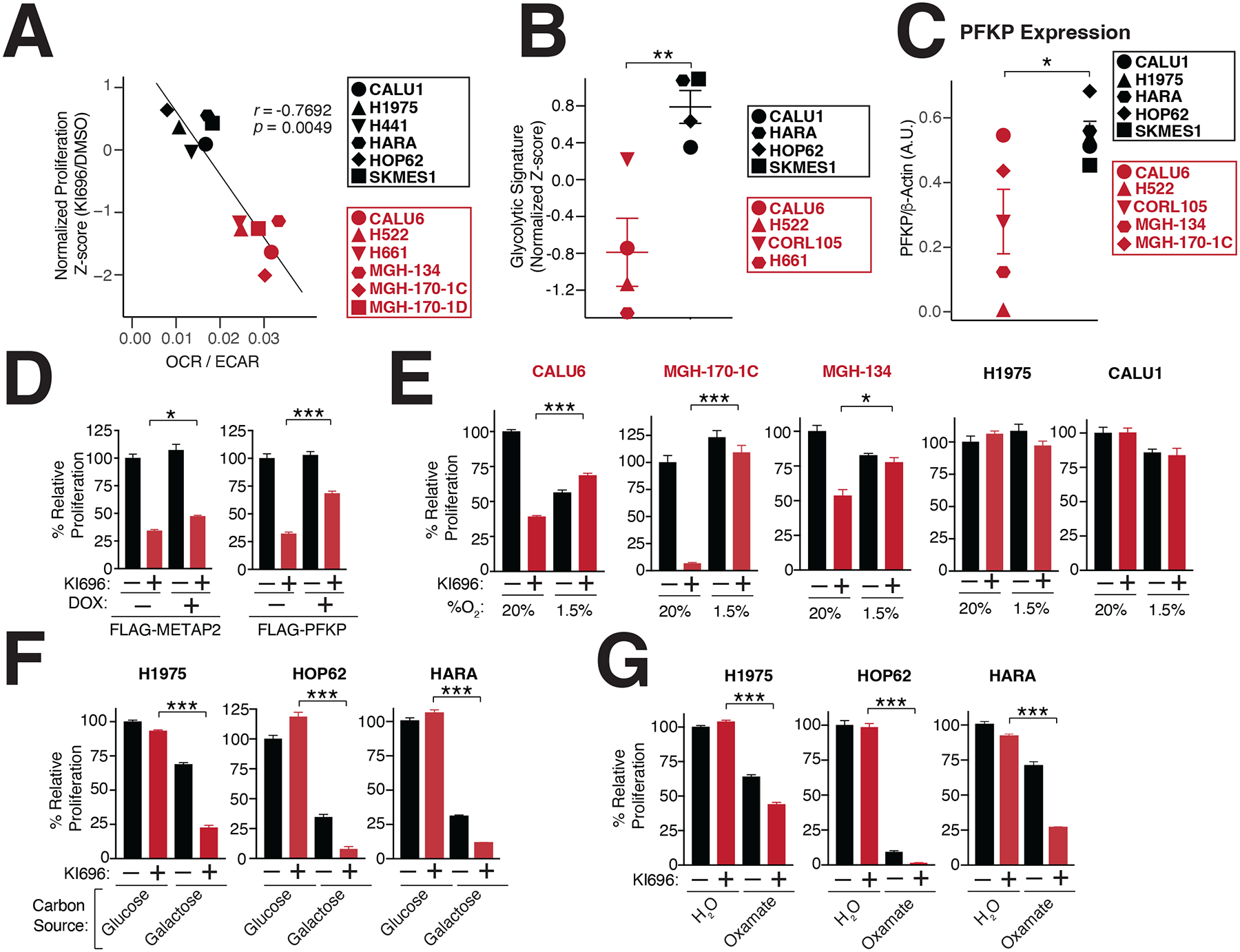
(A) NRF2 sensitivity correlates with higher levels of oxidative metabolism. Oxygen consumption rate (OCR) and extracellular acidification (ECAR) were measured in a panel of NSCLC cells and the OCR/ECAR for each cell line was plotted against its corresponding sensitivity to NRF2 activation (Data are represented as a mean ± SEM, n=6–8 biological replicates). (B) KEAP1-dependent cells have a lower glycolytic gene signature (see also Table S3). (C) The rate-limiting glycolytic enzyme phosphofructokinase, platelet isoform (PFKP), is highly expressed in KEAP1-dependent cells. Quantification of PFKP levels relative to β-actin (see also Figure S2G). (D) PFKP overexpression restores proliferation following NRF2 activation. Relative proliferation of CALU6 cells expressing FLAG-PFKP or FLAG-METAP2 (control) was determined by crystal violet staining following doxycycline (DOX) (100 nM) and KI696 (1 μM) treatment (Data are represented as a mean ± SEM, n=5 biological replicates). (E) Hypoxia rescues NRF2 sensitivity. Relative proliferation in a panel of NSCLC cell lines following treatment with KI696 (1 μM) and culture in normoxic (20% O2) or hypoxic conditions (1.5% O2) was determined as in (D) (Data are represented as a mean ± SEM, n= 5 biological replicates). (F-G) Glycolytic inhibition sensitizes cells to NRF2 activation. KEAP1-independent cells were treated with KI696 (1 μM) and cultured in media containing glucose (10 mM) or galactose (10 mM) (F) or co-treated with sodium oxamate (10 mM) (G) and relative proliferation was determined as in (D) (Data are represented as a mean ± SEM, n=5 biological replicates). (H) NRF2 activation decreases maximal respiration in KEAP1-dependent cells. Maximal respiration was determined in a panel of NSCLC lines following treatment with KI696 (1 μM) for 48 hrs (Data are represented as a mean ± SEM, n=6–8 biological replicates). * indicates p-values < 0.05, *** indicates p-values < 0.0001. One-way ANOVA with Sidak’s post-hoc correction and two-tailed student’s t-test were used to determine statistical significance.
