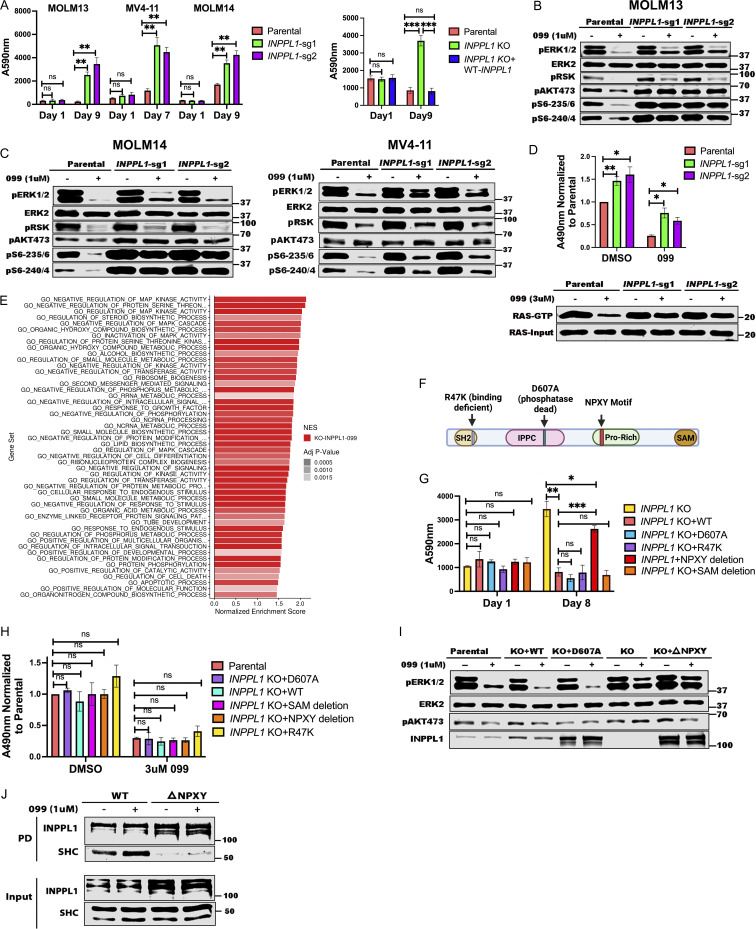
Figure 2.
INPPL1 KO acts downstream of RAS to promote ERK reactivation and SHP099 resistance. (A) INPPL1 KO causes resistance to SHP099. Left: PrestoBlue assays of MOLM13, MOLM14, and MV4-11 cells, treated with 1, 2, or 2 µM SHP099, respectively. Right: Proliferation of parental MOLM13 cells, INPPL1 KO MOLM13 cells, and INPPL1 KO MOLM13 cells re-expressing WT INPPL1 in 1 µM SHP099. (B and C) Immunoblots of lysates from parental and INPPL1 KO MOLM13 (B), MOLM14 (C), and MV4-11 (C) cells treated with vehicle or SHP099 (1 µM) for 1 h; note increased activity of ERK pathway (pERK, pS6-235/6) in SHP099-treated INPPL1 KO cells. (D) INPPL1 KO causes increased RAS activation in MOLM13 cells. Cells were treated with vehicle (DMSO) or SHP099 (3 µM) for 1 h, and RAS-GTP was quantified by active RAS ELISA (Cytoskeleton). Luminescence at A490 nm was normalized to parental DMSO values. Lower panel shows RAS RBD-pulldown assays on MOLM13 cell lysates. (E) Top 50 significantly different MSigDB GO biological processes in INPPL1 KO vs. parental MOLM13 cells based on differentially expressed genes in RNAseq. (F) Domain structure of INPPL1 indicating mutants/deletions of functional domains assessed. (G) NPXY motif but not lipid phosphatase activity of INPPL1 is required to restore SHP099 sensitivity. WT INPPL1 and the indicated INPPL1 mutants were re-expressed in INPPL1 KO MOLM13 cells, and cell number after exposure to SHP099 (1 µM) for 8 d was inferred by PrestoBlue assays. (H) NPXY motif is dispensable for INPPL1 effects on RAS activation. WT INPPL1 and the indicated INPPL1 mutants were re-expressed in INPPL1 KO MOLM13 cells. Cells were treated with vehicle (DMSO) or SHP099 (3 µM) for 1 h, and RAS activation was assessed by ELISA. All readings are normalized to vehicle-treated parental MOLM13 cells. (I) NPXY motif is required for ERK regulation by INPPL1. Parental or INPPL1 KO MOLM13 cells reconstituted with the indicated mutants were treated with SHP099 (1 µM) for 1 h, and cell lysates were prepared and analyzed by immunoblotting with the indicated antibodies. (J) NPXY motif is required for SHC binding. INPPL1 KO MOLM13 cells were reconstituted with WT- or NPXY deleted-INPPL1 bearing a C-terminal Strep tag. Cells were treated with vehicle (DMSO) or SHP099 (1 µM) for 1 h, and INPPL1 was recovered on Strep-Tactin beads and analyzed by immunoblotting as indicated. All immunoblotting experiments were independently performed three times except for those in J, which were performed twice. *, P < 0.05; **, P < 0.01; ***, P < 0.001. Source data are available for this figure: SourceData F2.