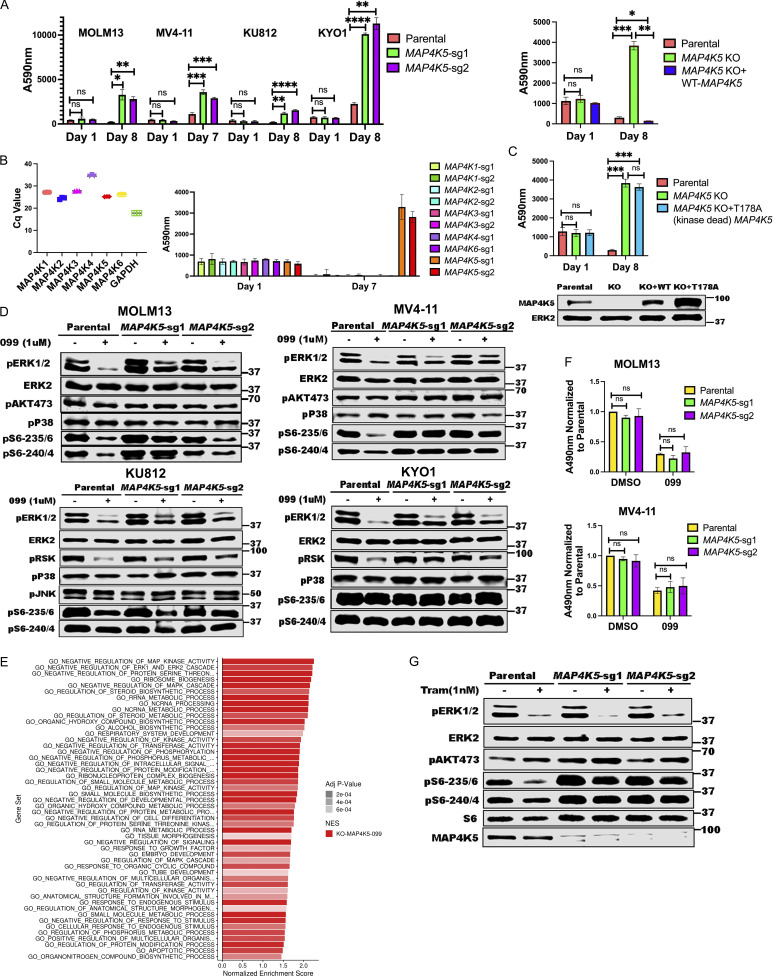
Figure 3.
MAP4K5 KO promotes SHP099 resistance downstream of RAS in a kinase-dependent manner. (A) MAP4K5 KO causes SHP099 resistance. Left: PrestoBlue assays on MOLM13, MV4-11, KU812, or KYO1 cells treated with 1, 2, 5, or 10 µM SHP099, respectively. Right: PrestoBlue assays of parental MOLM13 cells, MAP4K5 KO MOLM13 cells, and MAP4K5 KO MOLM13 cells reconstituted with WT MAP4K5 in the presence of SHP099 (1 µM). (B) Deletion of MAP4K5, but not other MAP4K family members, causes resistance to SHP099 in MOLM13 cells. Left: All MAP4K family members are expressed in MOLM13 cells as detected by qRT-PCR. Right: Only MAP4K5 KO causes resistance to SHP099. MOLM13 cells with deletion of the indicated MAP4K family member gene were treated with SHP099 (1 µM) and assessed by PrestoBlue assay on day 7. (C) MAP4K5 requires kinase activity to restore SHP099 sensitivity. Top: Proliferation assays on parental MOLM13 cells, MAP4K5 KO MOLM13 cells, or MAP4K5 KO MOLM13 cells reconstituted with kinase-dead MAP4K5 (T178A) in the presence of 1 µM SHP099. Bottom: Immunoblot showing reconstitution of MAP4K5 KO MOLM13 cells with WT or kinase-dead (T178A) MAP4K5. The immunoblotting experiment was performed once. (D) Increased ERK pathway activity in SHP099-treated MOLM13, MV4-11, KU812, and KYO1 cells lacking MAP4K5. Cells were treated with SHP099 (1 µM) for 1 h, lysed, and analyzed by immunoblotting with the indicated antibodies. Experiments for each cell line were independently performed twice. (E) Top 50 significantly different MSigDB GO biological processes in SHP099-treated MAP4K5 KO vs. parental MOLM13 cells, based on differentially expressed genes from RNAseq. (F) MAP4K5 KO does not affect RAS activation in MOLM13 or MV4-11 cells. MOLM13 and MV4-11 cells were treated with vehicle (DMSO) or SHP099 (3 µM MOLM13, 5 µM MV4-11) for 1 h, lysed, and assessed for RAS activation by ELISA. (G) MAP4K5 regulates ERK activation upstream of MEK. Parental or MAP4K5 KO MOLM13 cells were treated with vehicle (DMSO) or the MEK inhibitor trametinib (1 nM) for 1 h, lysed, and analyzed by immunoblotting. Experiments were independently performed three times. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. Source data are available for this figure: SourceData F3.