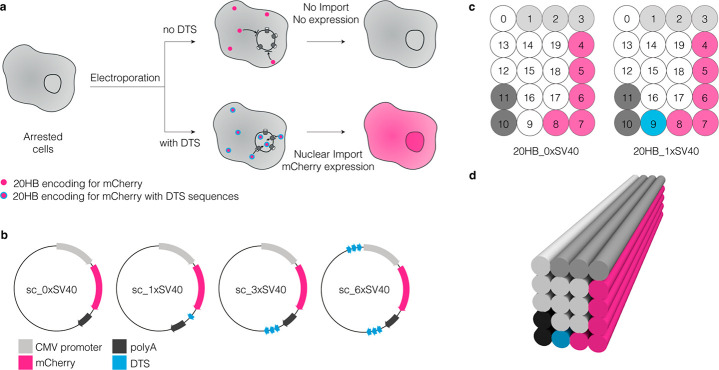
Figure 1.
Engineering nuclear localization signals into custom DNA origami scaffolds and structures. (a) Electroporation enables delivery of DNA origami directly to the cytoplasm. Cellular recognition of DTS sequences should enhance nuclear uptake and thus expression of mCherry in nondividing cells, which are modeled here using a chemically arrested cell model. (b) Plasmid designs with varying numbers of SV40 DTS sequences included (0×, 1×, 3×, and 6×SV40 repeats) for production of custom ssDNA scaffolds. (c) Schematic cross section of helices (0–19) for the 20HB structures displaying sequences of interest in the exterior helices. (d) Cylindrical model of the used DNA origami structure, a 20-helix bundle (20HB). The colored regions in (b), (c), and (d) display the gene encoded on the scaffold part in the respective helix.