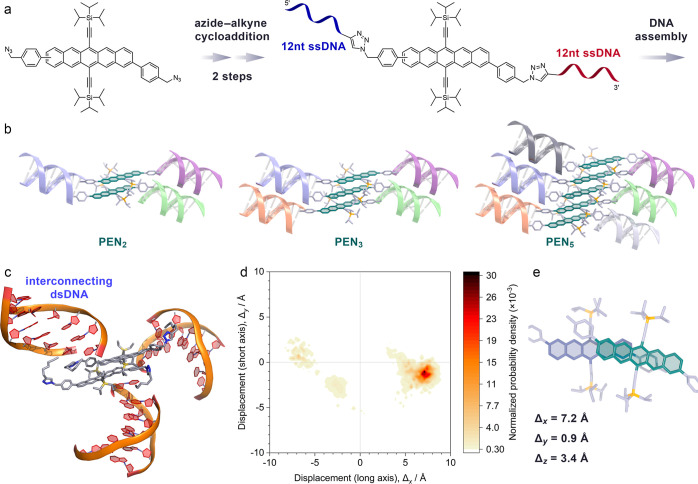
Figure 1.
Assembly and modeling of the pentacene/DNA constructs. (a) DNA-functionalized pentacenes are synthesized via Cu(I)-catalyzed azide–alkyne cycloaddition. Two 12 nucleotide (nt) single-stranded DNAs (ssDNAs) of different base sequences are attached sequentially while preserving the directionality of the DNA. This process is repeated to generate a library of pentacenes with complementary sequences. Pentacene dimers, trimers, and pentamers are generated by selecting the desired components from this library, followed by hybridization to form rigid dsDNAs. (b) Schematic illustration of the DNA-linked pentacene constructs (PENn) that consist of n pentacenes, interconnected by (n – 1) dsDNA and two terminating dsDNAs. Color coding represents complementary base sequences. (c) Simulated MD-optimized geometry of PEN2. The DNA arranges radially around the hydrophobic pentacenes. (d) Normalized probability density map showing the lateral offset along the long (x) and short (y) molecular axes of the pentacenes in PEN2. Driven by hydrophilic–hydrophobic interactions and guided by the local geometry due to the bulky TIPS-ethynyl substituents, the pentacenes are positioned in a well-defined arrangement. (e) Cut-out of the optimized PEN2 geometry, showing the arrangement of the pentacenes with considerable offset along the long molecular axis. The pentacenes are closely π-stacked with an average distance of 3.4 Å.