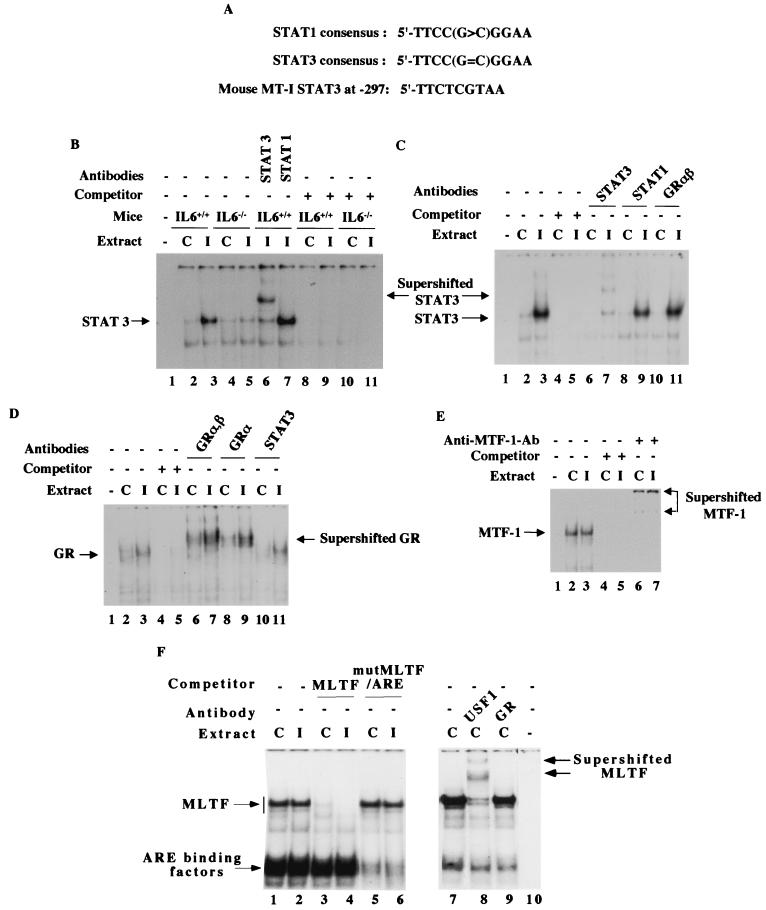
FIG. 6.
Analysis of DNA-binding activities of different transcription factors that bind to the MT-I promoter in the liver nuclear extract from virus-infected and control mice. (A) Homology of the IL-6 type II RE of mouse MT-I promoter to STAT3 and STAT1 consensus sites. (B) STAT3 but not STAT1 binds to IL-6 type II RE of the mouse MT-I promoter, which is significantly increased in the liver nuclear extract of influenza A virus-infected wild-type mice but not that of IL-6 knockout mice. Nuclear extracts (10 μg of protein) from each group of mice were incubated on ice for 30 min with 32P-labeled STAT3 oligonucleotide (0.1 to 0.2 ng) in the binding buffer [12 mM HEPES (pH 7.9), 75 mM KCl, 5 mM MgCl2, 0.5 mM DTT, and 2 μg of poly(dI-dC)]. The DNA-protein complexes were separated at 150 V for 4 h at room temperature on a polyacrylamide (6% acrylamide) gel with 0.5× TBE as the running buffer. The dried gel was subjected to autoradiography as well as the PhosphorImager. For supershift assay, the nuclear extract was incubated on ice with 0.4 ng of anti-STAT1 (sc-346) or anti-STAT3 (sc-483) antibodies (Santa Cruz Biotechnology) before addition of the labeled oligonucleotide. For competitive EMSA, the reaction mixture was incubated with a 100-fold molar excess of the unlabeled oligonucleotide before addition of the labeled probe. Lane 1 represents the reaction mixture without the extract. (C) Glucocorticoid receptor does not physically interact with STAT3–IL-6 type II RE complex formed on MT-I promoter. The liver nuclear extracts from the control (lanes C) and infected (lanes I) IL-6+/+ mice were incubated with either anti-STAT3, anti-STAT1, or anti-GRαβ (sc-1004) antibodies. For competitive EMSA, the reaction mixture was incubated with a 100-fold molar excess of the unlabeled oligonucleotide before addition of the labeled probe. (D) The DNA-binding activity of the glucocorticoid receptor (GR) is increased in the liver nuclear extract from infected mice. Nuclear extracts (20 μg of protein) were incubated with 32P-labeled GRE1 oligonucleotide (0.2 to 0.4 ng) in the binding buffer containing 12 mM HEPES (pH 7.9), 60 mM KCl, 5 mM MgCl2, 0.5 mM DTT, and 4 μg of poly(dI-dC) on ice for 30 min at 4°C. DNA-protein complexes were separated on a polyacrylamide gel (4% acrylamide) with 0.5× TBE as the running buffer. For supershift assay, the nuclear extract was incubated for 1 h with 0.2 μg of anti-GRαβ, GRα (sc-1002), or STAT3 IgG (Santa Cruz Biotechnology) before addition of the labeled oligonucleotide. Lane 1 indicates the labeled oligonucleotide incubated without any extract. For the competition experiment, the reaction mixture was incubated with a 100-fold molar excess of the unlabeled oligonucleotide before addition of the corresponding labeled oligonucleotide. (E) Constitutive DNA-binding activity of MTF-1 is high in the liver nuclear extract of wild-type and IL-6 null mice and is not increased in the extract from infected animals. Nuclear extracts (10 μg of protein) were incubated at 4°C for 20 min with 32P-labeled MRE-s oligonucleotide (0.2 to 0.4 ng), to which MTF-1 binds specifically, in the same binding buffer as that of STAT3. The DNA-protein complexes were separated on a polyacrylamide gel (4% acrylamide) with 0.25× TBE as the running buffer. For supershift assay, the reaction mixture after the binding reaction was incubated for 1 h with 1 μl of anti-MTF-1 antiserum. For competitive EMSA, the reaction mixture was incubated with a 100-fold molar excess of the unlabeled oligonucleotide before addition of the labeled probe. Lane 1 denotes the labeled oligonucleotide incubated in the absence of extract. (F) The DNA-binding activities of factors that bind to MLTF/ARE composite site are not altered in the liver nuclear extract from infected animals. Nuclear extracts (10 μg of protein) were incubated with 32P-labeled MLTF/ARE oligonucleotide following the same conditions as described for STAT3. The DNA-protein complexes were resolved by electrophoresis on a polyacrylamide gel (6% acrylamide) with 0.5× TBE as the running buffer. For supershift assay, the extract was incubated with anti-USF1 (sc-229) antibodies (Santa Cruz Biotechnology) in the binding buffer before the reaction. The DNA-protein complexes were analyzed by autoradiography and quantitated by Storm system (Amersham).