Lassnig et al. 10.1073/pnas.0408589102. |
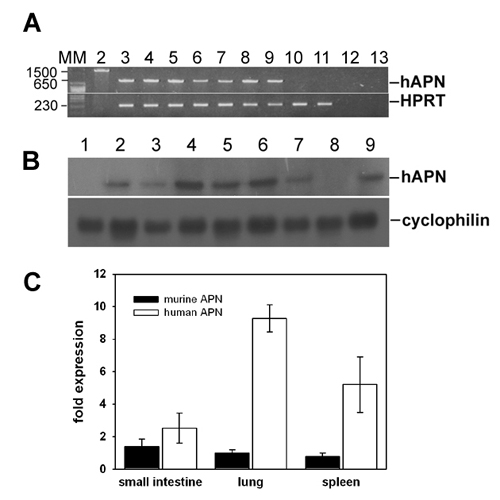
Fig. 5.
Expression of hAPN mRNA in transgenic mice. (A) RT-PCR to detect human APN transcripts in diverse organs. The transgene-specific 650-bp signal appeared in kidney, spleen, lung, liver, small intestine, heart, and brain (Upper, lanes 3-9, respectively) from transgenic animals. No transgenic RNA was detected in different tissues from nontransgenic animals (lung and kidney, lanes 10 and 11, respectively). The relative quantity of the cDNA was examined by amplifying in parallel the ubiquitously expressed hypoxanthine guanine phophoribosyl transferase (HPRT) (Lower). Lane 2, transgenic DNA; lane 12, minus-RT control; lane 13, nontemplate control. The length of the different PCR products in bp is indicated on the left. MM, molecular markers in bp. (B) Northern blot analysis of different transgenic lines. Total RNA (12 mg) isolated from the small intestine of different mice was fractionated on glyoxal/DMSO gels and analyzed by Northern blot hybridization. Transgene transcripts with the expected length were detected in the small intestine of all investigated mouse lines (Upper, lanes 2-7) and in the transfected murine epithelial cells (lane 9). In contrast, the same amount of RNA isolated from the small intestine of nontransgenic mice did not contain any human APN transcript (lanes 1 and 8). The blot was stripped and subsequently rehybridized with a cyclophilin-specific probe (Lower). (C) Real-time RT-PCR was performed to compare the endogenous (murine APN) with the transgenic APN (hAPN) level. The hAPN expression levels were normalized to HPRT and calibrated to endogenous APN expression in WT mice. hAPN expression levels were similar to endogenous APN levels in small intestine and spleen, whereas higher hAPN expression levels could be verified in the lung. In each experiment, 8-week-old WT (n = 8) and transgenic (n = 8) mice were analyzed in parallel. These data were confirmed by four additional independent experiments (n = 8 for each).
Supporting Materials and Methods
Total RNA was isolated from 1 × 107 cultured cells or 1 g of different tissues by using TRIzol reagent (Invitrogen). First-strand cDNA synthesis from 1 mg of total RNA was carried out by using oligo(dT) primers and Superscript II RNase H-Reverse Transcriptase (Invitrogen). Transgene mRNA expression was monitored by combining primers hAPNex19 and hubglobR (see Materials and Methods). For an amplification control, murine hypoxanthine guanine phophoribosyl transferase (HPRT) (GenBank accession no. NM013556) was analyzed with muHprt-forward 5'-TTG CTC GAG ATG TCA TGA AGG A-3' and muHprt-reverse 5'-TGA GAG ATC ATC TCC ACC AAT AAC TT-3' (nt 233-254 and nt 456-431, GenBank accession no. NM013556). Full-length transgene cDNA was amplified by using hAPN5'-forward 5'-CAT CAC CAT GGC CAA GGG-3' (nt 18-35, GenBank accession no. X13276) and hubglobR (see above).
Real-time RT-PCR quantification of APN transcripts was carried out as described in refs. 1 and 2 by using the following primers: muHprt-forward and muHprt-reverse (see above); APN-forward 5'-GAG TTC GAG CTG CAG CAG-3' and APN-reverse 5'-ATC TTT GTT CTC CTT CAC CCA-3'. TaqMan probes were 5'-FAM-TGG GAG GCC ATC ACA TTG TGG C-TAMRA-3' (muHprt), 5'-FAM-TAA AGC GGA TAA CTC AGC CAC AGG-TAMRA-3' (muAPN), and 5'-FAM-CTG GAG CAA GCC CTG GAG AAG AC-TAMRA-3' (hAPN).
Northern blot analysis was performed as described in ref. 3. 32P-labeled probes were a 650-bp hAPN::b-globin cDNA (RT-PCR: hAPNex19 and hubglobR; see Fig. 1A) and a mouse cyclophilin cDNA probe (nt 7-626, GenBank accession no. X52803) as endogenous control.
1. Pfaffl, M. W., Georgieva, T. M., Georgiev, I. P., Ontsouka, E., Hageleit, M. & Blum, J. W. (2002) Domest. Anim. Endocrinol. 22, 91-102.
2. Karaghiosoff, M., Steinborn, R., Kovarik, P., Kriegshauser, G., Baccarini, M., Donabauer, B., Reichart, U., Kolbe, T., Bogdan, C., Leanderson, T., et al. (2003) Nat. Immunol. 4, 471-477.
3. Muller, M., Laxton, C., Briscoe, J., Schindler, C., Improta, T., Darnell, J. E., Jr., Stark, G. R. & Kerr, I. M. (1993) EMBO J. 12, 4221-4228.