Chang et al. 10.1073/pnas.0504547102. |

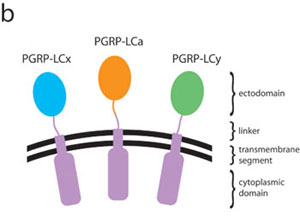
Fig. 6.
Sequence comparisons and topological arrangement of the ectodomains of peptidoglycan-recognition protein (PGRP)-LCa, -LCx, and –LCy. (a) Sequence alignment of the ectodomains of the three PGRP-LC isoforms; an aligned sequence of PGRP-SA is also shown at the bottom. Secondary-structure elements in the LCa ectodomain are indicated above the alignment. The residue number of the LCa ectodomain is also shown above the alignment. Insertions in the LCa ectodomain are highlighted in red. Invariant residues are boxed in dark red and colored in white; conserved residues are shaded in light blue. The disulfide bond-forming Cys residues are linked with a black line. The two nonconserved N-linked glycosylation sites in PGRP-LCa are in bold letters with the confirmed site marked with a black circle. (b) The N-terminal residues of the LCa, LCx, and LCy ectodomains are connected to identical transmembrane and intracellular domains via linkers, which consist of 40 residues in -LCa but are 20-residue shorter in both -LCx and -LCy. Such topological arrangement of the PGRP-LC receptor would allow the front face of the ectodomain to be accessible to other molecules in the extracellular milieu; on the other hand, the back side of the ectodomain is attached with the linker and thus could not be exploited for potential protein-protein/ligand interactions.
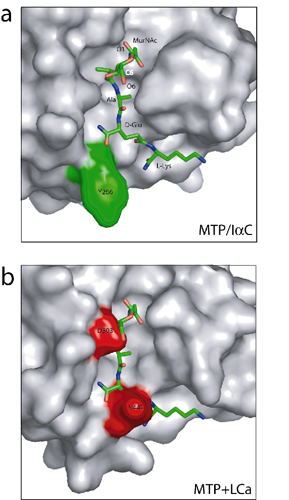
Fig. 7.
The LCa ectodomain structure would not allow a typical docking interaction with a muropeptide. (a) Closeup view of the structure of N-acetylmuramic acid (MurNAc)-l-Ala-d-Gln-l-Lys (MTP) complexed to PGRP-IaC. The MTP molecule is shown as a stick model. PGRP-IaC is in surface representation. Note that the bound MTP is oriented such that the C1-O1 and C6-O6 hydroxyl bonds are solvent-exposed. (b) MTP modeled onto the front face of LCa ectodomain by superimposing the latter with the MTP-PGRP-IaC structure; PGRP-IaC was omitted for clarity. PGRP-LCa residues Asn-442 and Asp-503 are highlighted in red.
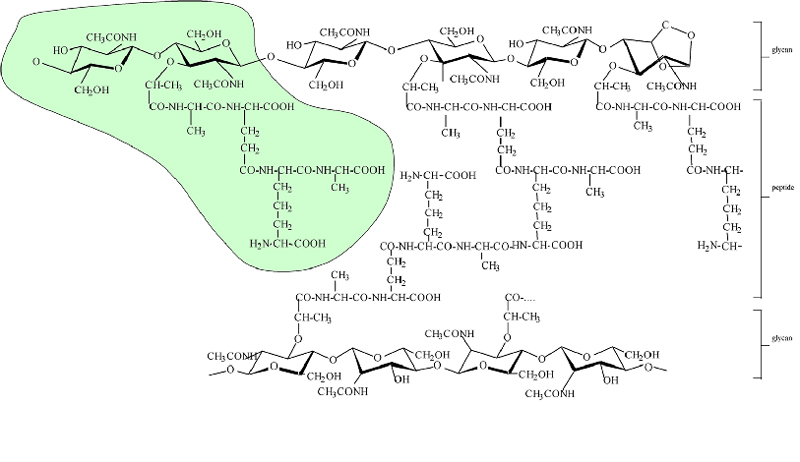
Fig. 8.
The chemical structure of diaminopimelic acid (DAP)-typed peptidoglycan in Escherichia coli. The structure of the highlighted muropeptide is GlcNAc-MurNAc-l-Ala-gamma-d-Glu-meso-DAP-d-Ala. The glycan strands ended naturally with a 1,6-anhydro MurNAc residue, which is shown on the right of the scheme. Note that the peptidoglycan structure varies from one bacterial species to another, mainly in the structure of the peptide moiety.Table 1. Data collection and refinement statistics
Wavelength, Å | 1.0000 |
Resolution, Å | 50-2.5 |
Space group | P4132 |
Unit cell parameters, Å | a = b = c = 105.08 |
No. of total reflections | 48848 |
No. of unique reflections | 7439 |
Redundancy (last shell) | 6.6 (6.6) |
Completeness (%)(last shell) | 99.4 (99.6) |
Intensities I/s I (last shell) | 27.4 (2.4) |
R merge* (%) (last shell) | 6.2 (55.8) |
R work/Rfree† (%) | 21.3/24.1 |
No. of atoms (protein) | 1316 |
No. of atoms (all) | 1365 |
rms deviation in bond length, Å | 0.010 |
rms deviation in bond angles, ° | 1.27 |
Average B values, Å2 | 51.7 |
Ramachandran plot statistics, %‡ | |
Residues in most favored region | 90.5 |
Residues in additionally allowed region | 8.8 |
Residues in disallowed region§ | 0.7 |
*Rmerge = S |(Ihkl) - < I> |/S (Ihkl), where Ihkl is the integrated intensity of a given reflection.
†
R = (S |Fo – Fc|)/(S Fo), where Fo and Fc are observed and calculated structure factors, respectively.‡
Calculated by using the program procheck [Laskowski, R. A., Rullmannn, J. A., MacArthur, M. W., Kaptein, R. & Thornton, J. M. (1996) J. Biomol. NMR 8,477-486].§
The y and f angles of Asp507 are in a disallowed region. However, this residue has well defined electron density.