Ludwig et al. 10.1073/pnas.0502954102. |

Fig. 5.
Defense responses and ethylene signaling triggered by the truncated VK(59K) variant of Nicotiana tabacum calcium-dependent protein kinase (NtCDPK)2. Leaves were infiltrated with Agrobacterium strains containing full-length [variable/protein kinase/junction/C-terminal calmodulin-like domain (VKJC)], truncated (VK59K) or truncated kinase-inactive (VKD/A) variants of NtCDPK2 or the Cf-9 disease resistance gene. At day 3, leaves were subjected to an abiotic stress stimulus (injection of water), and subsequent responses were analyzed. (A) Induction of hypersensitive reaction (HR)-like cell death 3 days after infiltration. (B) Western blot analysis of solubilized membrane extracts representing samples harvested at indicated time points after infiltration confirming the expression of full-length NtCDPK2 (VKJC), VK59K, and VKD/A. The active CDPK kinase variants show a mobility shift caused by posttranslational modifications (C). Synthesis of hydrogen peroxide upon infiltration with 3,3'-diaminobenzidine solution and forceps stimulus. (D) Northern blot analysis of total RNA isolated over a time course after an abiotic stress stimulus. Membranes were hybridized with HinI, PR1a, and PR1b probes and equal loading was confirmed by prior methylene-blue staining (MB). (E) Determination of 1-aminocyclopropane-1-carboxylate (ACC) synthesis. Pooled discs of three independent leaves per sample were harvested over a time course after an abiotic stress stimulus from control leaves (open circle), leaves expressing full-length NtCDPK2 (filled circle), VK59K (diamond), VKD/A (open square), or in a biotic gene-for-gene interaction after elicitation of Cf-9-expressing leaves with Avr9 (filled triangles) or control intercellular fluid (IF; open triangles). (F) Interference of CDPK and mitogen-activated protein kinase (MAPK) signaling in the Avr9/Cf-9 gene-for-gene interaction. Samples from leaves expressing Cf-9 or coexpressing NtCDPK2 variants VK59K and VKD/A were harvested before and 30 min after infiltration of Avr9. Protein expression was confirmed by Western blot analysis (Upper) and activation of MAPKs salicylic acid protein kinase (SIPK) and wound-induced protein kinase (WIPK) was analyzed by an in-gel kinase assay using myelin basic protein as substrate (Lower). (G) Avr9/Cf-9-dependent MAPK activation analyzed by an in-gel kinase assay as described for F, except that samples originate from VK-coexpressing leaves pretreated with 40 mM silver thiosulphate as indicated.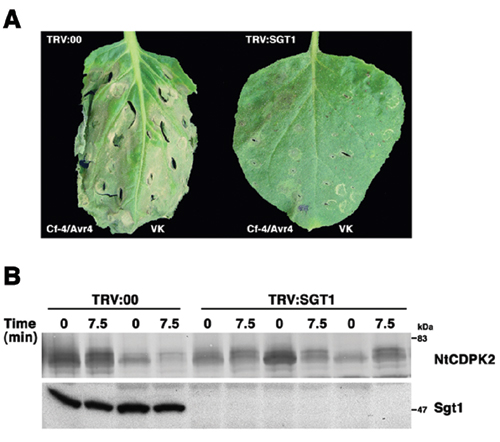
Fig. 6.
Nicotiana tabacum calcium-dependent protein kinase (NtCDPK)2-signaling dependent cell death but not NtCDPK2 activation requires SGT1. (A) Comparison of cell death response in wild-type and SGT1-silenced plants caused by the Avr4/Cf-4 gene-for-gene interaction or by NtCDPK2 variable domain/protein kinase domain (VK)-dependent signaling after an abiotic infiltration stress stimulus. Silencing of NbSGT1 was induced by inoculating Nicotiana benthamiana seedlings with Agrobacterium tumefaciens strains TRV:SGT1 or TRV:00 (control). After 3 weeks, both components of the Avr4/Cf-4 gene-for-gene interaction were transiently expressed in the left leaf halves, and VK was expressed in the right leaf halves, which were in addition subjected to the abiotic stress 3 days after the infiltration of Agrobacterium strains. The hypersensitive response in the gene-for-gene interaction (left half) was comparable with the NtCDPK2-signaling induced necrosis (right half) in nonsilenced plants (documented at day 4). No cell death response was observed on either leaf side in SGT1-silenced plants. (B) Protein expression of NtCDPK2-myc (VKJC) and SGT1 in wild-type and NbSGT1-silenced N. benthamiana plants. Three days after infiltration of agrobacteria strains, leaf discs were harvested from different leaves before and 7.5 min after a secondary stress stimulus. Silencing of SGT1 was confirmed by Western blot analysis of soluble extracts using an anti-SGT1 antibody (Lower), and SGT1-independent expression and activation (visible as a mobility shift) of NtCDPK2-myc protein was determined in solubilized membrane extracts using an anti-c-myc antibody (Upper). Irrespective of the silencing of SGT1, ectopically expressed full-length NtCDPK-myc still showed the stress-induced mobility shift 7.5 min after stimulation.Table 1. Agrobacterium tumefaciens GV3101 strains
Name | Purpose | Description of binary vector | Source |
Empty vector control | Transient expression | Empty pBIN19 vector | 1 |
VKJC | Transient expression | 35 S-NtCDPK2 full-length gene fused to C-terminal triple c-myc tag in pBIN19 | 2 |
VKJC(3) | Transient expression | 35 S-NtCDPK3 full length gene fused to C-terminal HA tag in pBIN61 | This study |
VK59K | Transient expression | 35 S truncated NtCDPK2, coding for variable and kinase domain only, fused to triple c-myc tag in pBIN19 | This study |
VK | Transient expression | 35 S truncated NtCDPK2, lysine 59 to glutamate point-mutation, fused to triple c-myc tag in pBIN19 | 2 |
VK(D/A) | Transient expression | 35 S truncated NtCDPK2, point-mutated in the catalytic aspartate residue to alanine (D241A), fused to triple c-myc tag in pBIN19 | This study |
VK(3) | Transient expression | 35 S truncated NtCDPK3, coding for variable and kinase domain only, fused to HA tag in pBIN61 | this study |
GG | Transient expression | 35 S double GFP gene in pBIN61 | This study |
J-GG | Transient expression | 35 S junction domain from NtCDPK2 fused with C-terminal double GFP in pBIN61 | This study |
Cf9:TAP | Transient expression | 35 S Cf-9 fused to C-teminal TAP tag in pBIN19 | 3 |
4/456/Avr4 | Hr | 35 S Cf-4, 35S Avr4 | 4 |
TRV:00 | Silencing | Empty TRV-vector | 5 |
TRV:SGT1 | Silencing | 634 bp of NbSGT1 inserted into TRV vector | 6 |
1. Bevan, M. (1984) Nucl. Acid Res. 12, 8711–8721.
2. Romeis, T., Ludwig, A. A., Martin, R. & Jones, J. D. G. (2001) EMBO J. 20, 5556–5567.
3. Rivas, S., Romeis, T. & Jones, J. D. G. (2002) Plant Cell 14, 689–702.
4. Thomas, C. M., Tang, S., Hammond-Kosack, K. & Jones, J. D. G. (2000) Mol. Plant-Microbe Interact. 13, 465–469.
5. Ratcliff, F., Martin-Hernandez, A. M. & Baulcombe, D. C. (2001) Plant J. 25, 237–245.
6. Peart, J. R., Lu, R., Sadanandom, A., Malcuit, I., Moffett, P., Brice, D. C., Schauser, L., Jaggard, D. A., Xiao, S., Coleman, M. J., et al. (2002) Proc. Natl. Acad. Sci. USA 99, 10865–10869.
Table 2. Primers used for Northern blot probes
Gene name | Primers sequences |
PR1aF | 5′-AGT AAT ATC CAC TCT TGC CG-3′ |
PR1aR | 5′-AGA ACA ACA TAT CCT CC-3′ |
PR1bF | 5′-AAG CTC AAA ACT CTC CCC-3′ |
PR1bR | 5′-CAT CAG TTG GAA GTT CCA AC-3′ |
PR2aF | 5′-GGG CAC AAT CTA TTG GAG-3′ |
PR2aR | 5′-TTG CCC TCT GAT CAG GAG-3′ |
PR2bF | 5′-GGG CTC AAT CGA TAG GTG-3′ |
PR2bR | 5′-TTT CAA CTG AAC TGT CCC-3′ |
Hin1F | 5′-ACT GAA ACT AGT ATG GCC CTT CCA TTC CGC-3′ |
Hin1R | 5′-ATT CTA CCA TGG CTA CCA ATC AAG ATG GCA-3′ |
tpoxF | 5′-AGT TTA CTA ATT GGA AGC TCA TCA-3′ |
tpoxR | 5′-TTC TCC ACA GCA GAT TTG ATA TTG-3′ |
PI-IIF | 5′-ATG GCT GTT CAC AAA GTT AGT TTC-3′ |
PI-IIR | 5′-AGC AGC ACT TTG AGG CTC CCC CAC C-3′ |
F, forward; R, reverse.
Supporting Materials and Methods
The tandem GFP fusion genes were generated by PCR with primers B-GFP-f (5'-CCC GGA TCC GGC ATG AGT AAA GGA GAA GAA CTT TTC-3') and GFP-Bg-r (5'-GGG ACT AGT TTA AGA TCT TTT GTA TAG TTC ATC CAT-3'), introducing a BamHI site at the 5' end and a BglII and SpeI site at the 3' end, and using 35 S promoter GFP construct as template. The J-moiety encoding fragment was amplified by PCR with primers Sl-J-f (5'-CCG CGT CGA CCC ATG GCA CCA GAT AAG CCT CTG G-3') and J-B-r (5'-GGC GGA TCC GAT GAC TCT CAA AGC CAT TTT CTT G-3') coding for an N-terminal SalI site and a C-terminal BamHI site. The J–GFP–GFP fusion construct was assembled as a SalI–J–BamHI/BamHI–GFP1–BglII/BamHI–GFP2–SpeI fragment, whereby the second GFP-encoding fragment was inserted as a BamHI/SpeI fragment into the BglII and SpeI sites of the first to generate GFP–GFP. Blunted SalI/SpeI or BamHI/SpeI fragments were inserted into the SmaI site of binary vector pBIN61, yielding plasmids SLJ20342 (J–GFP–GFP) or SLJ20341 (GFP–GFP). Plasmids were electroporated into Argobacterium tumefaciens GV3101.