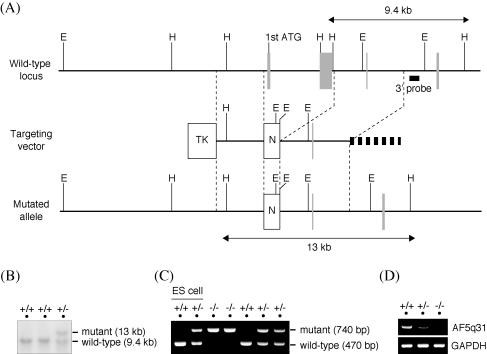
FIG. 2.
Targeted disruption of the AF5q31 gene. (A) Schematic representation of the wild-type allele of mouse AF5q31 (top), the targeting vector (middle), and the mutant allele resulting from a homologous recombination (bottom). Filled boxes are exons, and open boxes are selection marker genes. H, HindIII restriction site; E, EcoRI restriction site; N, neomycin resistance gene cassette; TK, thymidine kinase gene cassette. (B) Southern blot analysis of HindIII-digested genomic DNAs (5 μg/lane) from ES clones with an external 3′ probe. The 9.4-kb and 13-kb bands represent the wild-type and targeted alleles, respectively. An external 3′ probe used to analyze is shown in panel A. (C) PCR-based genotype analysis of tail DNAs isolated from the pups of AF5q31+/− intercrosses. Three kinds of primers (see Materials and Methods) detected both the wild-type allele (470-bp band) and the targeted allele (740-bp band). As controls, parental and targeted ES cells were used. (D) RT-PCR analysis of total RNAs from AF5q31+/+, AF5q31+/−, and AF5q31−/− MEFs. The primers located on exons I and IV of the AF5q31 gene were used. RT-PCR for GAPDH confirms equivalent amounts of RNAs used for the analysis.