Bogani et al. 10.1073/pnas.0500584102. |

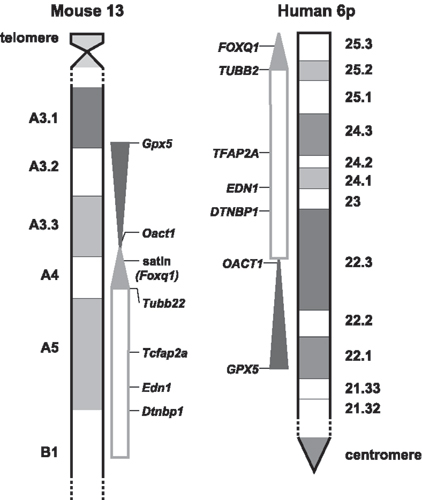
Fig. 4. Human and mouse synteny. Ideograms of proximal Mmu 13 and the short arm of human 6 with the extent of Del(13)Svea36H indicated by the adjacent shaded region. The nonshaded box represents the region conserved between Mmu13 and 6p, but not deleted in the Del(13)Svea36H mouse. Note the rearrangement of the genetic material such that on Mmu13 the 6p25, -24, and -23 analogous segment lies distal to material that corresponds to 6p22. Additionally, the 6p22 material is inverted on the mouse chromosome (represented by the direction of the dark shading).
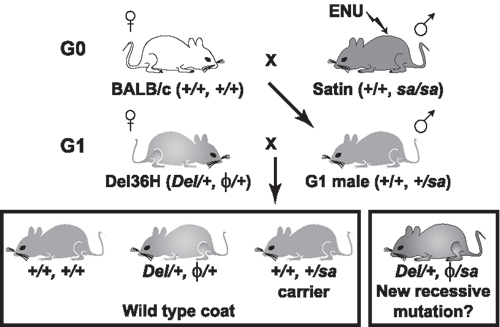
Fig. 5. A genetic screen at Del(13)Svea36H on mouse chromosome 13. The breeding protocol for the genetic screen is shown, with the f symbol used to indicate hemizygosity at the particular locus due to the presence of the Del(13)Svea36H chromosome.
Table 3. Polymorphic markers across Del(13)Svea36H
Marker | Position, Mb | C3H | 101 | BL/6 | B/c | Left primer | Right primer | PCR program |
D13Mit115 ¶ | 19.86 |
|
|
|
| TGGTGAAGTGTTTGGAAAAGG | TTTAACCCATTGATCTACTTCAAGG | 1 |
D13Mit17 ¶ | 20.67 |
|
|
|
| CACCCCCAAGTTCTCTTGAA | CCCACATACACATGTGCACA | 2 |
A34_298f22SNP2 § | 20.94 |
|
|
|
| ACGTTGGATGACTGGGCTGGTGTTAGATTG | ACGTTGGATGTGCTTCTTCCAGAAGACAGC | 5 |
A34_298f22SNP1 § | 20.94 |
|
|
|
| ACGTTGGATGAACCTCTAAGGGTGACACAG | ACGTTGGATGTGATCAAATGCTACTCAGGC | 5 |
DNR059 ¶ | 21.16 |
|
|
|
| GTTCTCACGGCCATTTCAG | CCATGATGTGAGCAAGAAACC | 3 |
13.019.659* | 21.78 |
|
|
|
| TCTTCTCTCTGAGTCATGCC | TGTCATGTGGGCACAGAAAG | 5 |
13.020.129* | 22.25 |
|
|
|
| GTTGGATGAAATCAGTTTCACAGTCTGC | GTTGGATGGGGCACAAAACCTCTTTCTC | 5 |
WI_WGS_13_22488197 † | 22.47 |
|
|
|
| ACGTTGGATGGTAATGTCTGGATCTGTGGG | ACGTTGGATGTCCTCCGACACTGTAAAAAC | 5 |
rs3679575 ‡ | 23.08 |
|
|
|
| ACGTTGGATGAGGGCAGCAAACCCCAATTC | ACGTTGGATGTGAAACTCACTTCCTCCCTG | 5 |
H46_480b19SNP § | 23.10 |
|
|
|
| ACGTTGGATGTCAGAAAAAGCCATGGGCTC | ACGTTGGATGTGCAGATGGAGCAAGAAGTG | 5 |
WI_WGS_13_23196359 † | 23.16 |
|
|
|
| ACGTTGGATGTCCAACCTTCAGACACCAAC | ACGTTGGATGTGGTTTCTCACCTGGTGAGG | 5 |
WI_WGS_13_23196389 † | 23.17 |
|
|
|
| ACGTTGGATGTTAGCCTCACCAGGTGAGAA | ACGTTGGATGTCCTTTACCATGCCTAGGAG | 5 |
WI_WGS_13_23196397 † | 23.17 |
|
|
|
| ACGTTGGATGTCCTTTACCATGCCTAGGAG | ACGTTGGATGTCACCAGGTGAGAAACCAAC | 5 |
WI_WGS_13_23199897 † | 23.17 |
|
|
|
| ACGTTGGATGTTGAGATAGTGTTTGGGTAG | ACGTTGGATGGCTGACATTGATCATGGAGC | 5 |
WI_WGS_13_23254757 † | 23.23 |
|
|
|
| ACGTTGGATGAGTAGAGCTATTGAGGTGGG | ACGTTGGATGGCAATTTCACACTGATGGCG | 5 |
WI_WGS_13_23254874 † | 23.23 |
|
|
|
| ACGTTGGATGTCAGCATGAAGCCTCAAAGC | ACGTTGGATGACACACGCAACTGGGAAAAC | 5 |
WI_WGS_13_23254887 † | 23.23 |
|
|
|
| ACGTTGGATGGAAGCCTCAAAGCATCTTCC | ACGTTGGATGACACGCACACACGCAACTGG | 5 |
WI_WGS_13_23255016 † | 23.23 |
|
|
|
| ACGTTGGATGCTGTTTCTGCTTCCTCAGTG | ACGTTGGATGCTTAAGAGCTCAGAACCCAC | 5 |
D13Mit14 ¶ | 23.78 |
|
|
|
| GGAACAGCAAGCTCTAAGGG | CTACCAGGCCTCCCAAGATA | 3 |
D13Mit15 ¶ | 23.78 |
|
|
|
| AGGAACAGCAAGCTCTAAGGG | GGCCTCCCAAGATATCATCA | 3 |
WI_WGS_13_24350948 † | 24.32 |
|
|
|
| ACGTTGGATGAACCCCGCGGCGCAGCTAC | ACGTTGGATGAAGCTGTCCCCGCGGAGTAG | 5 |
D13Mit84 ¶ | 25.00 |
|
|
|
| TATGTATCCACAGCCCTTAAAATG | TGAGTTGCATCCTTGTAACCC | 3 |
D13Mit133 ¶ | 25.61 |
|
|
|
| TAGACACTTAATTCTGTGATGAAATGG | AGCAAAAGCCCCAGTTAGTG | 3 |
DNR048 ¶ | 26.62 |
|
|
|
| CCCTACACACAAGGTTCAGG | CATGCAGTCAAAACATGCTG | 3 |
DNR076 ¶ | 27.67 |
|
|
|
| ACAGGAGCAGCTTGTCATTG | CATTAAGCTCCACCCATTCC | 3 |
DNR101 ¶ | 28.31 |
|
|
|
| CATTAAGCTCCACCCATTCC | GCAATTAGCAGGAAGGTGATG | 3 |
TETNR5 ¶ | 29.40 |
|
|
|
| TCTCCTGCATGCTCTGTGTC | AACAGCCTGGTCTATAATGCAAG | 3 |
13.027.892* | 30.06 |
|
|
|
| GTTGGATGTCCTTGTCTGTCCTTTGCTG | GTTGGATGGGAGAGCTTAAGAGGCATAC | 5 |
TETNR10 ¶ | 30.28 |
|
|
|
| GCTGATGCTAGGGCTTTCTC | CCCTAGGGCACATTTCACTG | 3 |
13.028.317* | 30.48 |
|
|
|
| GTTGGATGGTGGCAGTAGTAGACTGAAG | GTTGGATGTATGGGCCATTTCTTGCCTG | 5 |
rs3654710 ‡ | 30.80 |
|
|
|
| ACGTTGGATGTATGCTTAGAGCATGGGCAG | ACGTTGGATGCCCACATGCATGTTTATCCC | 5 |
13.028.654* | 30.82 |
|
|
|
| GTTGGATGCTACCAACTCCAGAGAGAAG | GTTGGATGGGCTCTTTATCCCTAGCTTC | 5 |
satin | 30.94 |
|
|
|
| GAGATCAACGAGTACCTCATGGG | CGAAGGAGCTGGAGAACTTG | 4 |
13.029.103* | 31.27 |
|
|
|
| GTTGGATGCCTGGTACTCATCAAAGGTC | GTTGGATGGGTGTTCATGGTACAGACAG | 5 |
kkSNP04 § | 31.43 |
|
|
|
| ACGTTGGATGCAGACCAGATGCACAAAAAG | ACGTTGGATGCCTGCCTTTTAGTCCACAAC | 5 |
13.029.431* | 31.60 |
|
|
|
| GTTGGATGAGATGGTCTAATATTAGCAC | GTTGGATGTGAGGCCAACACAAACAATG | 5 |
rs3687359 ‡ | 31.79 |
|
|
|
| ACGTTGGATGGTGGTGCTTTGCTAACTGTG | ACGTTGGATGGGACATGTTTGGTTTCCAAG | 5 |
DNR024 ¶ | 31.94 |
|
|
|
| AGCCCACTTCAACTCCTCAG | TTCTGCACCCATATTTTCATTG | 3 |
13.029.993* | 32.16 |
|
|
|
| AGGACTCTGCAGTAAGAGAC | TGGCATTTGAGAGGGAAGAC | 5 |
13.030.240* | 32.41 |
|
|
|
| GTTGGATGTGCCTTTGTGGATGAGTTGC | GTTGGATGAGCAGGACCATCAAGCTCAG | 5 |
rs3023380 ‡ | 32.43 |
|
|
|
| ACGTTGGATGCACTCCCTAATCTCTGGAAC | ACGTTGGATGCCTCAGCAAAATCTTGCAAAC | 5 |
DNR03 ¶ | 32.76 |
|
|
|
| GGAGAAAACAGACTTCATTCAGG | AAGTCAAACATGGCTAGTG | 3 |
13.031.633* | 32.93 |
|
|
|
| GTTGGATGGTATATTGTTTGTGTGTGGG | GTTGGATGTACCTCTGAGAGCATCAAAC | 5 |
DNR02 ¶ | 32.96 |
|
|
|
| ATCAACCTGGGCAAAGACTG | TTCTGCACCCATATTTTCATTG | 3 |
DNR016 ¶ | 33.33 |
|
|
|
| GGAGAATTGTGAGGGAATGC | CAGCCATCACATACCACACC | 3 |
D13Mit136 ¶ | 33.33 |
|
|
|
| TTTTATCTATTGAGTAGATTCATGGTG | TATGCCTGGAGGAAAACAGG | 1 |
13.032.078* | 33.38 |
|
|
|
| CAATGGCTTGCCAGATACAG | ATCCTCTCCATTTCTGTCCC | 5 |
D13Mit275 ¶ | 36.76 |
|
|
|
| TTAGCAAGGGAACAGAGAGAGG | CAATCAAGGTATCCCTGTCTCC | 2 |
1. PCR conditions: 95°C for 15 min, followed by 94°C for 1 min, 55°C for 30 s, 72°C for 45 s for 35 cycles; and then 72°C for 5 min.
2. PCR conditions: 95°C for 15 min, followed by 94°C for 1 min, 56°C for 30 s, 72°C for 45 s for 35 cycles; and then 72°C for 5 min.
3. PCR conditions: 95°C for 15 min, followed by 94°C for 1 min, 64.5°C for 30 s, 72°C for 45 s for 14 cycles; and then 95°C for 45 s, 57° C for 30 s, 72°C for 30 s for 30 cycles, 72°C for 5 min.
4. PCR conditions: 95°C for 15 min, followed by 94°C for 30 s, 55°C for 35 s, 72°C for 30 s for 35 cycles and then 72°C for 5 min, in the presence of 5 M Betaine.
5. PCR conditions: 95°C for 1 min, followed by 94°C for 20 s, 56°C for 30 s, 72°C for 60 s for 45 cycles; and then 72°C for 3 min.
* Pletcher M. T., McClurg P., Batalov S., Su A. I., Barnes S. W., Lagler E., Korstanje R., Wang X., Nusskern D., Bogue M. A., et al. (2004). PLoS Biol 2, e393.
†
Petkov P. M., Ding Y., Cassell M. A., Zhang W., Wagner G., Sargent E. E., Asquith S., Crew V., Johnson K. A., Robinson P., et al. (2004) Genome Res 14, 1806‡
http://www.ncbi.nlm.nih.gov/projects/SNP/§
SNPs were discovered with Homogenous Mass Cleave SNP discovery technique [Stanssens, P., Zabeau, M., Meersseman, G., Remes, G.,Gansemans, Y., Storm, N., Hartmer, R., Honisch, C., Rodi, C. P., Bocker, S. et al. (2004) Genome Res 14, 126-133.] All SNPs assays were performed by using MassARRAY [Bonora, E., Beyer, K. S., Lamb, J. A., Parr, J. R., Klauck, S. M., Benner, A., Paolucci, M., Abbott, A., Ragoussis, I., Poustka, A., et al. (2003) Mol. Psychiatry 8, 885-892.]¶
SSLP markers detected by fluorescent capillary electrophoresis using either the MegaBACE system (Amersham Biosciences) or the AB3100 Prism system.Table 4. Mutation detection screening
Fragment | Ensembl position | Region covered | Forward primer | Reverse primer | Detection method | PCR program |
Sox4 1 | 28332869 | 5’UTR/ Exon 1 | GAGCACTTCAGCGGTGA | GGAGATCTCGGCGTTGT | Sequencing | 12 |
Sox4 2 | 28332462 | Exon 1 | GCAAGATCATGGAGCAGTC | GAGAACGATGCAGCCG | Sequencing | 12 |
Sox4 3 | 28332151 | Exon 1 | GCCCAAGAAGAGCTGTG | GCGTAGGCTGGCGTA | Sequencing | 13 |
Sox4 4 | 28331823 | Exon 1 | GTTGTACGAAGATGGAGGC | GAATTCGAAGTGGGAGCCTG | Sequencing | 14 |
Sox4 5 | 28331604 | Exon 1/ 3’UTR | GACGACGAGTTCGAAGACGAC | CTCCTCTCCTGCCTCTTG | Sequencing | 15 |
Foxq1 1 | 30937553 | 5’UTR/ Exon 1 | CAACCAGTCCCTCGGCCTAAG | TTCAGCCTCGAGAGCCCATAG | Sequencing and DHPLC | 10 |
Foxq1 2 | 30938104 | Exon 1 | GGGCTCTCGAGGCTGAAGAC | GACAGTGGAGATGGCACGTC | Sequencing and DHPLC | 10 |
Foxq1 3 | 30938594 | Exon 1 | CCGGCATGAAATTGGAGGTG | AGGTTGTGGCGCACGGAGTT | Sequencing and DHPLC | 10 |
Foxq1 4 | 30939047 | Exon 1 | CCTTTTTCCGGGGCAGCTAC | CGGGAAGGAGCGCGGGATAG | Sequencing and DHPLC | 10 |
Foxq1 5 | 30939455 | Exon 1 | CGGCTCTGGGGGTGCAGCTA | GAGCCCACATTCCAACCTCCTC | Sequencing and DHPLC | 10 |
Foxq1 6 | 30939773 | Exon 1/ 3’ UTR | CCTACCCGGTGGAGACTCTG | ACACGCAGCAGTCACTCCTG | Sequencing and DHPLC | 10 |
Foxf2 1 | 31005686 | 5’UTR/ Exon 1 | CTCGCCCGATTTGTGCAC | AGCGCGATGTACGAGTAAGG | Sequencing | 18 |
Foxf2 2 | 31005762 | Exon 1 | AGTGGAGGCACCAAGAAGG | GGAACGAACCCTCCTCAAAC | Sequencing | 17 |
Foxf2 3 | 31006988 | Exon 1 | TTCCCCTTTTTCCGTGGCGC | TGGCCATATAGGTGGAGCCC | Sequencing | 16 |
Foxf2 4 | 31010164 | Exon1/ Intron 1 | TCAAGGCGGTTATGGTGGCC | AGAGGCTCTCAGAGGCTCCG | Sequencing | 17 |
Foxf2 5 | 31010328 | Intron 1/ Exon 2/ 3’ UTR | AGCTGCCTTTACACCCTCAG | ACAGTGTGAGTCCGTTGCAG | Sequencing | 17 |
Foxc1 1 | 31186196 | 5’UTR/ Exon 1 | CAGCCGAGTTCCTGAAGGACAG | CCTCGCAGCCCACTCAGTTC | Sequencing | 6 |
Foxc1 2 | 31186620 | Exon 1 | GCGTCCTGGTCTGGCCCTCTC | GGCGGCTTCACCATGTCCTTG | Sequencing and DHPLC | 7 |
Foxc1 3 | 31187025 | Exon 1 | GTGTACTCGCACCCTGCTCAC | TCCGTCTTGATGTCCTGGATG | Sequencing and DHPLC | 8 |
Foxc1 4 | 31187450 | Exon 1 | CCGGCTGCACCTCCAAGAACC | GAACTGCCCGCACTGGAGCTCT | Sequencing and DHPLC | 8 |
Foxc1 5 | 31187943 | Exon 1 | GCCTACTCTCCGGGCCAGAG | ATTCACCGGGGAGTTGTTCAAG | Sequencing and DHPLC | 9 |
Foxc1 6 | 31188411 | Exon 1 | CGGGAAATGTTCGAGTCTCA | CGAGTCTCTGAACGCAAGAA | Sequencing and DHPLC | 10 |
Foxc1 7 | 31188825 | Exon 1 | TCATGGTTTATTAAAGGACA | GACAGAGACTAGGGAACTC | Sequencing and DHPLC | 11 |
Foxc1 8 | 31189229 | Exon 1 | TAATAAATTGCCATTCAGTTTGA | TCCTTCTGTGTAATGCATAAAGA | Sequencing and DHPLC | 11 |
Foxc1 9 | 31189641 | Exon 1 | AACAGATGGAGATCAGCCTA | GGCAGATCACCCTAAGATAAT | Sequencing and DHPLC | 10 |
Foxc1 10 | 31189988 | Exon 1/ 3’ UTR | AGTCTGGGGTGGTTTC | GAATCAGAGGCCACGTAAGG | DHPLC | 10 |
Foxq1 1 | 30937553 | 5’UTR/ Exon 1 | CAACCAGTCCCTCGGCCTAAG | TTCAGCCTCGAGAGCCCATAG | Sequencing and DHPLC | 10 |
Foxq1 2 | 30938104 | Exon 1 | GGGCTCTCGAGGCTGAAGAC | GACAGTGGAGATGGCACGTC | Sequencing and DHPLC | 10 |
Foxq1 3 | 30938594 | Exon 1 | CCGGCATGAAATTGGAGGTG | AGGTTGTGGCGCACGGAGTT | Sequencing and DHPLC | 10 |
Foxq1 4 | 30939047 | Exon 1 | CCTTTTTCCGGGGCAGCTAC | CGGGAAGGAGCGCGGGATAG | Sequencing and DHPLC | 10 |
Foxq1 5 | 30939455 | Exon 1 | CGGCTCTGGGGGTGCAGCTA | GAGCCCACATTCCAACCTCCTC | Sequencing and DHPLC | 10 |
Foxq1 6 | 30939773 | Exon 1/ 3’ UTR | CCTACCCGGTGGAGACTCTG | ACACGCAGCAGTCACTCCTG | Sequencing and DHPLC | 10 |
6. PCR conditions: 95°C for 10 min, followed by 95°C for 30 s, 68.5°C* for 30 s, 72°C for 30 s for 14 cycles; then 95°C for 30 s, 61°C for 30 s, 72°C for 30 s for 25 cycles, 72°C for 5 min. (Using Qiagen reagents, gel excised PCR product and sub cloned with TOPO cloning kit).
7. PCR conditions: 95°C for 15 min, followed by 95°C for 30 s, 65°C for 30 s, 72°C for 30 sec for 30 cycles, 72°C for 5 min.
8. PCR conditions: 95°C for 10 min, followed by 95°C for 30 s, 67.5°C* 30 s, 72°C for 30 sec for 14 cycles; then 95°C for 30 sec, 60°C for 30 s, 72°C for 30 sec for 25 cycles, 72°C for 5 min.
9. PCR conditions: 95°C for 10 min, followed by 95°C for 30 s, 63.5°C* 30 s, 72°C for 30 sec for 14 cycles; then 95°C for 30 sec, 56°C for 30 s, 72°C for 30 sec for 25 cycles, 72°C for 5 min.
10. PCR conditions: 95°C for 10 min, followed by 95°C for 30 s, 70°C* 30 s, 72°C for 30 sec for 14 cycles; then 95°C for 30 sec, 62.5°C for 30 s, 72°C for 30 sec for 25 cycles, 72°C for 5 min.
11. PCR conditions: 95°C for 10 min, followed by 95°C for 30 s, 64.5°C* 30 s, 72°C for 30 sec for 14 cycles; then 95°C for 30 sec, 57°C for 30 s, 72°C for 30 sec for 25 cycles, 72°C for 5 min.
12. PCR conditions: 95°C for 10 min, followed by 95°C for 30 s, 59°C* 1 min, 72°C for 1 min for 14 cycles and then 95°C for 30 s, 52°C for 1 min, 72°C for 1 min for 32 cycles and then 72°C for 5 min in the presence of 10% DMSO and 1.5mM MgCl2.
Supporting Text
Mouse Strains and the Genetic Screen.
The satin (sa) mutant (1) was recovered from the Harwell embryo bank as a linkage stock also homozygous for the vestigial tail (vt) and muted (mu) alleles, maintained on a mixed C3H/HeH and 101/HeH background. The vt and mu alleles were segregated by crosses with the C3H/HeH strain and a closed colony of sa/sa mice segregating C3H/HeH and 101/HeH alleles was produced.Within the genetic screen, a new mutation was defined as follows. Based on the frequency of transmission of the deleted chromosome (2), amongst 25 progeny, five with a satin coat and 20 with a wild-type (agouti) coat are expected. To screen for visible phenotypes, mice with the satin coat were assessed weekly (until weaning) for the presence of dysmorphologies. Pedigrees with £1/25 satin progeny were considered to harbor a recessive lethal mutation and breeding was continued in these pedigrees until 50 progeny had been obtained, and the data reassessed by using a limit of £3/50 satin progeny. Defining a lethal as £3/50 satin progeny means that only six false positives should be encountered among every 1,000 pedigrees and generates a probability that 5% of lethal mutations (not of total pedigrees) will be missed because 4/50 satin progeny have been produced through recombination. The efficiency of ENU means that more than one mutation may be present in a single pedigree. If we assume an average mutagenesis frequency (1 × 10-3) (3) and a Poisson distribution, the likelihood of the Del(13)Svea36H segment containing two mutations is extremely low (3.8 × 10-9).
Neurological and Behavioral Assays.
The tests performed on all mice were Open Field Activity, a modified SHIRPA test, measurement of Grip Strength, Spontaneous Alternation in a Y-maze, Acoustic Startle Response with Prepulse Inhibition (PPI) and assessment of Swimming Ability. Grip strength was measured with an automated apparatus (Bioseb) using all four limbs of the test animal, and data presented as an average of five peak measurements (in grams). For the PPI assay, the startle responses were recorded in arbitrary units using a 110 dB, 40 ms white noise sound pulse ± a preceding 80 dB, 10-ms white noise prepulse and PPI calculated by expressing the prepulse + pulse response as a percentage of the pulse only response. Parameters in the Open Field Activity tests were measured using a TruScan system over 30 min in 10 3-min bins. Centre time represents the amount of time (in seconds) an animal spends in the central zone of the arena (equivalent to 16% of the total area). For all tests, ANOVA was used for the comparison between wild-type and mutant animals across different lines according to their behavioural evaluations. The ANOVA was followed by the Newman-Keuls test for post hoc comparisons and a Repeated Measures Analysis was performed to investigate statistical significance in multiple time measurement parameters.