West et al. 10.1073/pnas.0507360102. |

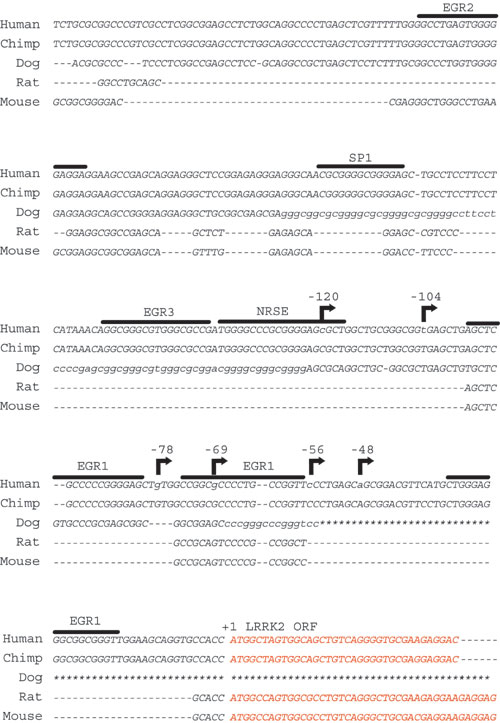
Fig. 7. Evolutionary conservation and structure of the predicted LRRK2 promoter. Genomic sequence from chimp, dog, or rodent is aligned to the human LRRK2-predicted promoter region by using a genome alignment tool (http://genome.ucsc.edu/cgi-bin/hgGateway). Positions of transcription start sites as determined in human brain are indicated with an arrow. Consensus recognition sequences for the indicated selected transcription factor in the predicted LRRK2 promoter are overlined, as determined by MATINSPECTOR. Red nucleotides indicate LRRK2 protein coding sequence. An * in the dog genome indicates an unsequenced region of the genome.
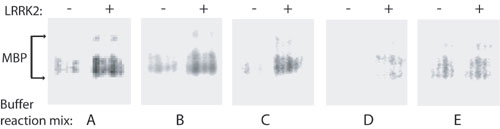
Fig. 8. Optimization of LRRK2 kinase activity. In vitro kinase activity of recombinant WT LRRK2 derived from transfected HEK-293T cells was assessed by using myelin basic protein as substrate. Incorporated 32P in myelin basic protein was resolved by using SDS/PAGE and imaged on a Storm phosphorimager. All reactions contain equivalent amounts of recombinant LRRK2, radioisotope, and myelin basic protein. Buffer A contains 10 mM imidazole (pH 7.5), 10 mM b-glycerol phosphate, 1 mM EGTA, 20 mM MgCl2, and 0.1 mg/ml BSA. Buffer B contains 20 mM Mops (pH 7.4), 25 mM b-glycerol phosphate, 1 mM EGTA, and 1 mM dithiothreitol. Buffer C contains 20 mM Hepes (pH 7.4), 15 mM MgCl2, 5 mM EGTA, and 20 mM b-glycerol phosphate. Buffer D contains 50 mM Hepes (pH 7.4) and 20 mM MgCl2. Buffer E contains 20 mM Tris-HCl (pH 7.4), 10 mM b-glycerol phosphate, 5 mM MgCl2, 2 mM MnCl2, and 0.5 mM dithiothreitol.