
Chen et al. 10.1073/pnas.0506618103. |

Fig. 6. gp78 and Ube2g2 are required for efficient degradation of CD3-d. HT1080 cells were transfected as in Fig. 1A. (A) Each of the three transfection conditions were split into three samples 24 h before treating for the indicated times with cycloheximide followed by resolution on SDS/PAGE and immunoblotting as indicated. The actin loading control is a reblot of the same membrane used to detect CD3-d. (B) HT1080 cells were labeled with 35S methionine/cysteine for 45 min followed by a chase of 3 or 6 h in complete medium. Anti-HA immunoprecipitates from cell lysates were resolved by SDS/PAGE.
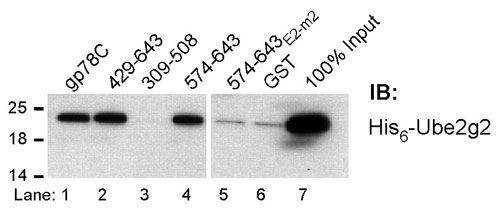
Fig. 7. gp78 directly binds Ube2g2 independent of its RING finger. Bacterially expressed His6-Ube2g2 was incubated with the indicated GST fusion proteins followed by immunoblotting (IB) with anti-His6.
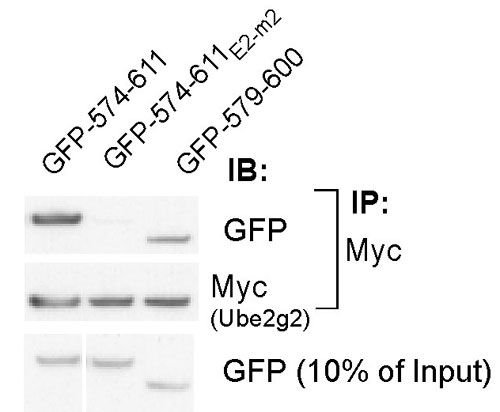
Fig. 8. The minimal G2BR binds Ube2g2 in cells. HEK 293T cells were transfected with GFP fusions together with plasmid encoding Myc-tagged Ube2g2 and IP carried out with anti-Myc (Top). Immunoblotting of IPs and 10% of input (Bottom) was with either anti-GFP (Top and Bottom) or with anti-Myc (Middle).
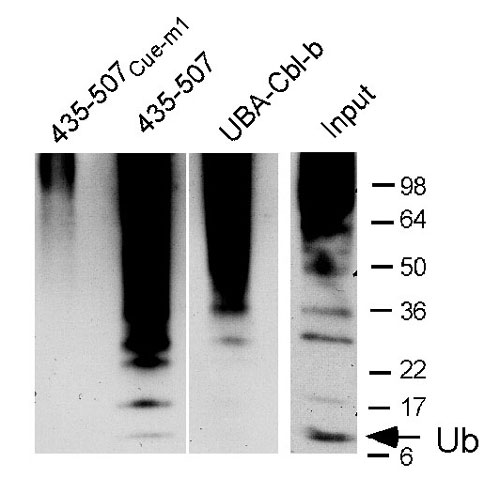
Fig. 9. Binding of ubiquitylated cellular species by the gp78 Cue domain. Equal amounts of cell lysates from cells treated with proteasome inhibitor (MG132) were incubated with the indicated GST fusions. After washing, samples were eluted by boiling, resolved by SDS/PAGE, transferred to membranes, and immunoblotted with antiubiquitin. Input lane is the same cell lysate used in the other lanes immunoblotted with the identical antibody but processed separately. Ladder of immunoreactive species below the 36-kDa marker corresponds to monoubiquitin (arrow) and predicted migration of ubiquitin oligomers. Note the relative prevalence of monoubiquitin in the lysate compared with that bound by the GST fusion of the gp78 Cue domain (435-507).

Fig. 10. p97/VCP binds the C terminus of gp78 independent of the G2BR. (A) Bacterially expressed p97 was incubated with the indicated GST fusion proteins of gp78 and binding assessed by IB with anti-p97. (B) HEK 293T cells were transfected as indicated and treated with MG132 for the final 8 h before harvesting. Lysates were immunoprecipitated with anti-gp78 followed by immunoblotting with anti-p97 or gp78 as indicated. (Lower) Equal levels of p97 in lysates from each of the transfections are demonstrated.

Fig. 11. Expression of gp78 mutants results in accumulation of CD3-d in the endoplasmic reticulum (ER) membrane. Membrane and soluble fractions from HEK 293T cells transfected as indicated were prepared as in Fig. 5A.
Table 1. Comparison of Ube2g2 and p97 binding
gp78 | Binds Ube2g2 | Binds p97 | Degrades CD3-d |
Full length | + | + | + |
611 truncation | + | - | + |
595 truncation | - | - | - |
E2-m2 mutation | - | + | - |
Rf-m | + | + | - |
Summary of coimmunoprecipitation results from Figs. 2E and 10B and CD3-d degradation results from Fig. 4 B and C.
Supporting Text
Cells and Antibodies
HT1080 [American Type Culture Collection (ATCC) CCL-121], human embryonic kidney (HEK) 293T (ATCC CRL-11268), and NIH 3T3 (ATCC CRL-1658) were maintained in DMEM with 10% FBS. Anti-Myc (9E10) and Flag (F3165) antibodies were from Sigma. Anti-GFP (sc-9996), actin (sc-1616), His6 (sc-803), and ubiquitin (sc-9133) were from Santa Cruz Biotechnology. Rabbit antisera against gp78 (1), UbcH5B, and ubiquitin (2), as well as monoclonal T cell antigen receptor (TCR)-a antibodies H28-710 (3) and A2B4-2 (4) have been described. Rabbit antisera for Ube2g2, Ube2g1, and hHRD1 were raised against peptides corresponding to 148-165, 160-170, and 493-509, respectively. Monoclonal anti-p97/VCP was a generous gift from Dr. Chou Chi Li (National Cancer Institute, Frederick, MD).
Plasmids
Plasmids encoding the following have been described: rat Ubc7 (Ube2g1) (5); UbcH5B (6); Myc-MmUbc7, C89S Myc-MmUbc7, HA-CD3-d, and 2B4 TCR-a (7); full length gp78 and the double RING finger mutation of gp78Rf-m in pCINeo (1); GST-UBA-Cbl-b (8); and p97/VCP (9). Constructs encoding His6-MmUbc7 in pCDNA-DEST47, GST- gp78 fusions in pGEX4T, and N-terminal Flag-tagged gp78 fragments in pFlag-CMV6 were generated by PCR. GFP-tagged constructs in pEGFP-C1 were generated by subcloning. Short-hairpin RNAs (shRNAs) against sequences in human AMFR/gp78 correspond to coding sequence bases 636-656 (gp78-sh1) and 318-336 (gp78-sh2), and Ube2g2 shRNA corresponds to bases 391-411 (Ube2g2-si1). Oligonucleotides were cloned into pSuper (OligoEngine, Seattle). Negative control shRNA provided with pSilencer (Ambion, Austin, TX) was subcloned into pSuper.
Binding Assays
GST fusions were purified from Escherichia coli. Binding was carried out by using equimolar amounts of fusion proteins measured by Coomassie blue. Generally, 20 picomole per sample were used. In vitro translated E2 was generated by coupled transcription and translation by using 35S methionine in either wheat germ or rabbit reticulocyte lysate (Promega, Madison, WI). Binding assays using in vitro translated E2 was carried out by incubating GST protein prebound to Glutathione-Sepharose 4B (Amersham Pharmacia Biosciences) with 105 cpm of 35S methionine labeled E2 in binding buffer (25 mM Tris·HCl, pH 7.4/50 mM NaCl/5 mM DTT/0.5% Nonidet P-40) overnight at 4°C. Beads were washed with binding buffer and resolved by SDS/PAGE under reducing conditions. Direct binding assays using purified bacterially expressed WT His6-MmUbc7 (His6-Ube2g2) and GST fusions were carried out by using the binding and wash conditions used for binding of in vitro translated E2s. For assessing thiolester formation and binding, Ube2g2 was cleaved and purified from GST-Ube2g2 and reacted with E1 and ubiquitin (6). Ube2g2 was then incubated with bead-bound GST fusions in the binding buffer described above without DTT followed by washing in the same buffer before elution and SDS/PAGE. Immunoblotting of Ube2g2 was carried out after transfer to Hybond-P (Amersham Pharmacia Biosciences) by using either anti-His6 or anti-Ube2g2 followed by ECL (Pierce). Tetraubiquitin binding studies were performed with binding and washing as above by using 1 mg of K48 tetraubiquitin (10). Binding of ubiquitylated material from cell lysates was carried out by using GST fusions that had been crosslinked to glutathione-Sepharose by using disuccinimidyl suberate (Pierce). Lysates were from HT1080 cells that were treated with 50 mM MG132 for 8 h.
Transfection Studies
Transfections were carried out »24 h after plating by using Polyfect (Qiagen, Valencia, CA) and cells harvested after »48 h. Vector without insert was used to equalize total plasmid. pEGFP-C1 served as an internal control. For experiments not involving immunoprecipitation, 6 × 105 cells were plated in six-well plates, and cell lysis was with Mammalian Protein Extraction Reagent (Pierce). Supernatants from a 13,000 × g centrifugation were used for further analysis. For immunoprecipitation (IP) of CD3-d or gp78 alone, 2.4 × 106 cells were seeded in 100-mm dishes, and cells were lysed in RIPA buffer (1 ´ PBS/1% Nonidet P-40/0.5% sodium deoxycholate/0.1% SDS) followed by IP from the supernatant of a 13,000 × g centrifugation and washing of IPs in RIPA buffer. For co-IP, cells transfected as above were lysed at 4°C in 50 mM Tris at pH 7.5/150 mM sodium chloride/1% Triton X-100/10% glycerol and supernatants from a 13,000 × g centrifugation analyzed. IPs were washed in the identical buffer used for lysis. All lysis buffers contained Protease Inhibitor Mixture (Sigma), and 10 mM iodoacetamide. MG132 (Sigma) was used at 50 mM. Cycloheximide was used at 50 mg/ml. For subcellular fractionation experiments, cell were resuspended in 0.25 M sucrose/10 mM triethanolamine/1 mM EDTA, pH7.4, for 10 min at 4°C followed by 15 passages through a 27-gauge needle. The product was subject to two sequential 5-min 1,000 × g spins to remove unbroken cells and nuclei. For separation of cytosolic and membrane fractions, supernatants from the 1,000 × g spin were pelleted by centrifugation at 100,000 × g for 1 h at 4°C. High-salt washes of membranes were carried out by resuspending pellets from the 100,000 × g centrifugation in 0.65 M NaCl/PBS after a 100,000 × g 1-h spin. Pellets and TCA-precipitated supernatants (cytosolic fraction) were analyzed. For metabolic labeling experiments, 36 h posttransfection, cells were incubated for 45 min in methionine-free medium and then labeled for 1 h with 200 mCi/ml of [35S]methionine/cysteine (ICN). Cells were then washed twice with complete medium followed by incubation in complete medium at 37°C. Cells were harvested at indicated times, and samples immunoprecipitated with anti-hemagglutinin (HA) followed by SDS/PAGE and imaging. For each time point, total counts used for immunoprecipitation were found to be within 10% of each other.
1. Fang, S., Ferrone, M., Yang, C., Jensen, J. P., Tiwari, S. & Weissman, A. M. (2001) Proc. Natl. Acad. Sci. USA 98, 14422-14427.
2. Lorick, K. L., Jensen, J. P., Fang, S., Ong, A. M., Hatakeyama, S. & Weissman, A. M. (1999) Proc. Natl. Acad. Sci. USA 96, 11364-11369.
3. Takata, M., Maiti, P. K., Kubo, R. T., Chen, Y. H., Holford-Strevens, V., Rector, E. S. & Sehon, A. H. (1990) J. Immunol. 145, 2846-2853.
4. Samelson, L. E., Germain, R. N. & Schwartz, R. H. (1983) Proc. Natl. Acad. Sci. USA 80, 6972-6976.
5. Lin, H. & Wing, S. S. (1999) J. Biol. Chem. 274, 14685-14691.
6. Jensen, J. P., Bates, P. W., Yang, M., Vierstra, R. D. & Weissman, A. M. (1995) J. Biol. Chem. 270, 30408-30414.
7. Tiwari, S. & Weissman, A. M. (2001) J. Biol. Chem. 276, 16193-16200.
8. Davies, G. C., Ettenberg, S. A., Coats, A. O., Mussante, M., Ravichandran, S., Collins, J., Nau, M. M. & Lipkowitz, S. (2004) Oncogene 23, 7104-7115.
9. Dai, R. M. & Li, C. C. (2001) Nat. Cell Biol. 3, 740-744.
10. Piotrowski, J., Beal, R., Hoffman, L., Wilkinson, K. D., Cohen, R. E. & Pickart, C. M. (1997) J. Biol. Chem. 272, 23712-23721.