
Barocchi et al. 10.1073/pnas.0511017103. |

Fig. 8. FACS analysis using RrgB antibodies of the invasive T4 strain (A), the clinical isolate of type 19F, ST16219F (B), the deletion mutant of mgrA, T4D(mgrA) (C), and the nonpiliated mutant T4D(rrgA-srtD), lacking the rlrA islet (D), showing the proportion of piliated versus nonpiliated cells. Respectively, 84%, 90%, 99%, and 0% of the bacteria express pili.
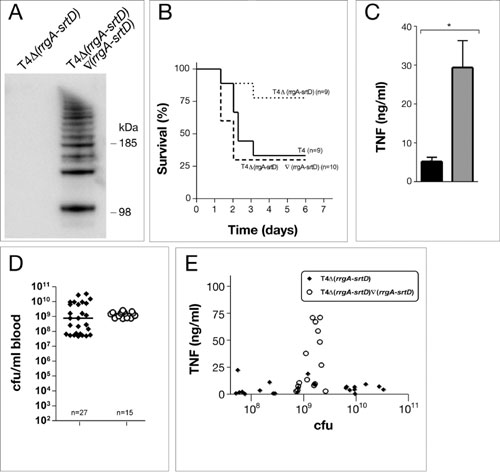
Fig. 9. (A) Western blot analysis with RrgB antibodies, on a 4-12% gradient gel, detects HMW polymers in the mutant where the rlrA islet has been reintroduced into the deletion mutant T4D(rrgA-srtD). (B) Survival of mice after intranasal inoculation with 5 × 106 (high dose) of the revertant where the rlrA islet has been reintroduced into the deletion mutant T4D(rrgA-srtD), as compared with T4D(rrgA-srtD) and T4. Virulence is restored to the same levels as for T4 having the rlrA islet. (C) The inflammatory response, after i.p. inoculation, is restored in the revertant where the rlrA islet has been reintroduced into the deletion mutant T4D(rrgA-srtD) (Mann-Whitney U test; *, P < 0.001). (D) Bacterial outgrowth in blood after i.p. challenge of mice with T4 and the revertant T4D(rrgA-srtD)Ñ(rrgA-srtD). (E) TNF response for individual mice correlated to the bacteremia levels after inoculation with T4 and the revertant T4D(rrgA-srtD)Ñ(rrgA-srtD).

Fig. 10. (A) Adherence of T4 and the mutant lacking rlrA (T4DrlrA) to monolayers of A549 lung epithelial cells. (B and C) Immunofluorescence microscopy of T4 (B), and T4D(rlrA) (C) adhering to A549 lung epithelial cells. Labeling of pneumococci with anti-capsular antibody (green) and visualization of epithelial F-actin with rhodamine (red) are shown. (D) Intranasal challenge of C57BL/6 mice with low/medium dose inoculation of T4 and T4D(rrgA-srtD), respectively, in separate experiments. Colonizations of individual mice are shown. The figure is a compilation of three different experiments.
Table 2. Primers and restriction enzymes used for creation of mutants
Gene | Name | Restriction enzyme | Sequence (5' to 3') |
erm cassette | ErmF | ApaI | TTTTTGGGCCCTTCGTGTTCGTGCTGACTTGC |
ErmR | BamHI | TTTTTGGATCCGATGTTGCTGATTAAGACGAGC | |
ErmstartR | AACTTCTTTTACGTTTCCGCC | ||
ErmslutF | ACCGAAAGACAGACGAGCC | ||
Kan-rpsL cassette | Kan-Bam | BamHI | TTGGATCCCTTTAAATACTGTAGAAAAGAGGA |
Kan-Apa | ApaI | TTGGGCCCTAAAACAATTCATCCAGTAAAAT | |
Dam406 | BamHI | TCTATGCCTATTCCAGAGGAAATGGATCGGATC | |
Dam351 | ApaI | CTAGGGCCCTTTCCTTATGCTTTTGGAC | |
Dam407 | AGGAGACATTCCTTCCGTATCT | ||
Dam352 | CAAGAGCACAGCGTGGTGCT | ||
T4D(rrgA-srtD) | RrgA P1 | CAAGGTCCAAACCTACTGAAC | |
RrgA P2 | ApaI | GCGGGCCCCTGAGATATACAGCACAGTCC | |
SrtD P3 | BamHI | CGGGATCCCTGGCATTTCTGGGAATCCTG | |
SrtD P4 | CGTTTCAAGTGCTATCACTGTTC | ||
T4D(mgrA) | MgrA P1 | ATATAACATGAACAGTTGGGTTCTTG | |
MgrA P2 | ApaI | ATATAGGGCCCAACCTCTTGCAATTATACCACA | |
MgrA P3 | BamHI | ATATAGGATCCCGCGTTTGAACTGTACCTCAA | |
MgrA P4 | ATATACAGTAACTGTCTCATCCAAATC | ||
MgrA C1 | ATATACTGCTTCAATCCATTAGTTATTTC | ||
MgrA C2 | ATATATTGATTGTAAAAATTCCATCTATAG | ||
T4D(rrgA-srtD)Ñ(rrgA-srtD) | Rev1 | BamHI | TTGGATCCTTATTTCCCTCGTAGTAAACGT |
Rev2 | ApaI | TTGGGCCCAAAGAAATGAAAGGAAAGCTAAGG | |
ST16219FD (rrgA-srtD) | RrgA P1 | CAAGGTCCAAACCTACTGAAC | |
RrgA P2 | ApaI | GCGGGCCCCTGAGATATACAGCACAGTCC | |
SrtD P3 | BamHI | CGGGATCCCTGGCATTTCTGGGAATCCTG | |
SrtD P4 | CGTTTCAAGTGCTATCACTGTTC | ||
SrD C2 | GCCCCATCTTGCCCTCACTGCG | ||
ST16219FD (mgrA) | MgrA P1 | ATATAACATGAACAGTTGGGTTCTTG | |
MgrA P2 | ApaI | ATATAGGGCCCAACCTCTTGCAATTATACCACA | |
MgrA P3 | BamHI | ATATAGGATCCCGCGTTTGAACTGTACCTCAA | |
MgrA P4 | ATATACAGTAACTGTCTCATCCAAATC | ||
MgrA C1 | ATATACTGCTTCAATCCATTAGTTATTTC | ||
MgrA C2 | ATATATTGATTGTAAAAATTCCATCTATAG | ||
D39Ñ(rrgA-srt)D(rlrA) | RLRAFR | CGCGGATCCAAAGGAGAATCATCATGCTAAACAAATACATTGA | |
RLRARX | CCCTCTAGATTATAACAAATAGTGAGCCTT |
Supporting Methods
FACS Analysis. Streptococcus pneumoniae [T4, ST16219F, T4D (mgrA), and T4D(rrgA-srtD)] isolates were grown in THY liquid culture overnight at 37°C under 5% CO2. Samples were diluted and allowed to grow to OD600 = 0.250 (»1 × 108 per ml). Bacterial cultures were centrifuged at 3,000 rpm and resuspended in 1× PBS. Fifteen microliters of bacterial suspension was added to 96-well plates. Five microliters of 20% normal rabbit serum was added to each well, along with primary antibodies (anti-RrgB, pre-immune anti-RrgB, and negative control Neisseria meningitidis anti-961) at 1:3,200. Samples were incubated on ice for 30 min, after which 150 ml of blocking buffer (1% BSA/PBS) was added to the wells. The 96-well plate was centrifuged at 2,500 rpm for 5 min at 4°C. A secondary anti-mouse antibody labeled with phycoerythrin (Jackson ImmunoResearch) was added at a final concentration of 1:100, and the mixture was incubated for 30 min on ice. Then 150 ml of blocking buffer was added and samples were centrifuged as above. Samples were resuspended in 200 ml of 1% paraformaldehyde/PBS and analyzed on the FACSCaliber (Becton Dickinson).
Creation of Revertant in T4D(rrgA-srtD).
The rlrA islet was reintroduced into T4D(rrgA-srtD) by reintroducing the knocked-out genes together with a kanamycin cassette. The kanamycin cassette was first integrated downstream of the target genes in the wild-type T4 strain by PCR ligation mutagenesis. Chromosomal DNA from these mutants was used to transform the knockouts and restore the wild-type phenotype. In the first step, the kanamycin cassette was amplified from Janus (1) with the primers Kan-Apa and Kan-Bam, creating a PCR product with ApaI and BamHI termini. Fragments upstream and downstream of the target sites were amplified with primers pili-rev-1-4 for the T4D(rrgA-srtD) mutant. The upstream fragments were constructed with ApaI sites and the downstream fragments with BamHI sites. All fragments were digested with corresponding restriction endonucleases and ligations were performed with equal molar amounts of upstream fragment, downstream fragment, and kanamycin fragment. After transformation into T4, transformants were selected on blood agar plates containing 200 mg/liter kanamycin. After control PCRs confirming the correct construction, chromosomal DNA from these transformants was used to transform T4D(rrgA-srtD). Again, selection was done on kanamycin-containing plates and the mutants were confirmed by checking for erythromycin sensitivity and pili expression.1. Sung, C. K., Li, H., Claverys, J. P. & Morrison, D. A. (2001) Appl. Environ. Microbiol. 67, 5190-5196.