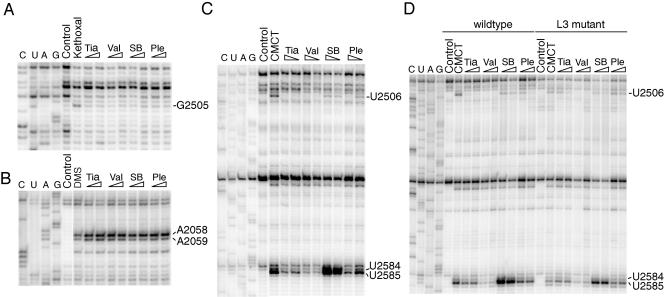
FIG. 2.
(A to C) Gel autoradiograms showing the antibiotic footprints on MRE600 ribosomes modified with (A) kethoxal, (B) DMS, and (C) CMCT. (D) Comparison of CMCT footprints on CN2476 (wild-type) and JB5 (L3 mutant) ribosomes. Nucleotides exhibiting altered reactivities in the presence of the pleuromutilin derivatives are indicated. Dideoxy sequencing lanes are designated G, A, U, and C. Lanes are labeled to indicate reactions with chemically unmodified 70S ribosomes in the absence of drugs (control) and 70S ribosomes modified in the absence of drugs (kethoxal, DMS, and CMCT). Ribosomes modified in the presence of tiamulin (Tia), valnemulin (Val), SB-264128 (SB), or pleuromutilin (Ple) are marked, where wedges are used to indicate a low (2 μM) or high (20 μM) drug concentration.