Jiang et al. 10.1073/pnas.0603804103. |
Supporting Figure 5
Supporting Figure 6
Supporting Figure 7
Supporting Movie 1
Supporting Materials and Methods

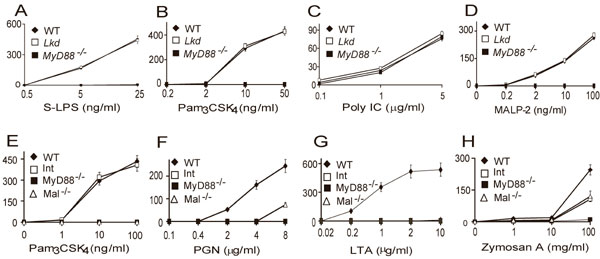
Fig. 5. The Lkd and Int phenotypes. Macrophages were isolated and treated as in Fig. 1. The inducers included smooth LPS (A), Pam3CSK4 (B and E), Poly I:C (C), MALP-2 (D), PGN (F), LTA (G), and zymosan A (H). TNF concentrations were assayed using the L929 bioassay.

Fig. 6. Ligand-dependent phosphorylation of signaling proteins and degradation of IkB in cells from germ-line mutant mice. Macrophages from WT, Poc (A), Lkd (B), and Int (C) mice were isolated and treated as indicated for different times. Macrophages were lysed and analyzed by immunoblotting with antibodies as indicated. (D) Mouse embryonic fibroblasts (MEFs) from heterozygous (Poc/WT), homozygous Poc (Poc), WT, and homozygous Lkd embryos were treated with IL-1 for the indicated times. Cells were then lysed and analyzed by immunoblotting with antibody against IkB and b-tubulin (loading control).

Fig. 7. Coarse genetic mapping of Lkd and sequencing of three mutant genes. (A) MyD88 mRNA was sequenced by using template from WT mice (Lower) or Poc homozygotes. A missense error (I179N) caused by a T®A transversion was found in Poc. (B) WT, MyD88-/-, and MyD88Poc/- macrophages (from Poc/Poc x MyD88-/- F1 hybrids) were isolated and treated with Pam3CSK4, CpG DNA, resiquimod, or Poly I:C. TNF production was assayed by bioassay. (C) Expression plasmids encoding WT-MyD88 or MyD88Poc were transfected into MEFs derived from Poc homozygotes together with an NF-kB reportor construct (pNiFty-luc). After 24 h cells were left untreated or treated with 100 units/ml IL-1 for 4 h, after which induction of luciferase was measured. Vector DNA (pcDNA3.1) was cotransfected into these cells together with pNiFty-luc as a control. (D) Coarse genetic mapping of Lkd. Phenotypic classification of F3 mice was based on a comparison of resiquimod-induced dose-response curves in which TNF production was measured by using the L929 bioassay. On 60 meioses, Lkd was confined to distal chromosome 9 with a peak LOD score of 17.16 (associated with D9Mit355). (E) MyD88 mRNA was sequenced by using template from WT mice (Lower) or Lkd homozygotes. A missense error (Y116C) caused by an A®G transition was found in Lkd. (F) Macrophages from WT mice, MyD88-/- mice, and MyD88Lkd/- mice (MyD88Lkd/Lkd ´ MyD88-/- F1 hybrids) were isolated and treated with resiquimod, and TNF production was assayed by bioassay. (G) TLR6 mRNA was sequenced by using template from WT (Lower) or Int homozygotes. A missense error (V327A) caused by an A®G transversion was found in Int.
Movie 1. Wire and ribbon rendering of association between the TIR domain of TLR2 (green) and MyD88 (cyan) predicted by docking studies. The BB loop of the receptor TIR domain with its critical proline residue and the Poc site valine are colored orange. The BB loop of the MyD88 TIR domain, with its critical proline residue and the Poc site isoleucine, are colored red.
Supporting Materials and Methods
Genetic Mapping and Positional Identification of Pococurante (Poc) and Lackdaisical (Lkd). Mutant homozygotes were outcrossed to C3H/HeN mice and backcrossed to the mutant stock. Mice were genotyped at 60 informative microsatellite loci. The mutation was confined between the two chromosome 18 markers (separated from the proximal marker by a single crossover and from the distal marker by numerous crossovers). Genotyping was performed by fragment length analysis using fluorescent primers and an ABI 3100 DNA sequencer. Sequence analysis was also performed with this machine and in all instances was performed on uncloned DNA fragments using internal primers.
Reagents.
Salmonella typhimurium (smooth) LPS, lipid A from Salmonella minnesota R595 (Re) (ultra pure, liquid), and MALP-2 were obtained from Alexis. Peptidoglycan was purchased from Fluka. Highly purified lipoteichoic acid was the kind gift of T. Hartung (Department of Biochemische Pharmakologie, Universität Konstanz, Germany). Unmethylated DNA oligonucleotide 5'-TCCATGACGTTCCTGATGCT-3' was synthesized by Integrated DNA Technologies (Coralville, IA). dsRNA was obtained from Amersham Pharmacia Biotech. Resiquimod was obtained from 3M. Pam2CSK4 and Pam3CSK4 were obtained from EMC Microcollections (Tübingen, Germany). Zymosan A was obtained from Sigma. Recombinant murine IFN-g, recombinant murine IFN-b, and IFN-a, IFN-g ELISA kits were obtained from R & D Systems. Antibodies against phosphorylated or total IkB, p38, c-Jun N-terminal kinase, and extracellular signal-regulated kinase 1/2 (p42/p44) were from Cell Signaling Technology (Beverly, MA), and antibody against HA was from Upstate Biotechnology(Charlottesville, VA). Antibody against b-tubulin was from Zymed (South San Francisco, CA).Bacterial Strain.
Streptococcus pyogenes Xen20 (derived from serotype M49, strain 591) was obtained from Xenogen (Alameda, CA). Listeria monocytogenes strain 10403S was made bioluminescent by transformation with the plasmid pAUL-A Tn4001 luxABCDE Kmr (1). L. monocytogenes strain 2C (flaA::lux/kan 10403S) was used as a donor for transducing luminescence. Transductions were performed as described (2) by using filtered supernatants of U153 or P35 phage-infected cultures of the donor strain added to logarithmic cultures of the recipient. The bacteria were selected for virulence by a passage in a C57BL/6 mouse.Infection with S. pyogenes or L. monocytogenes.
Six- to 10-week-old male and female mice were used. Hair was removed by chemical depilation, and the mice were injected s.c. with 100 ml of a midexponential growth phase (D600 = 0.6, 5 × 106 colony forming units ml-1) of S. pyogenes Xen20 resuspended in PBS and complexed to Cytodex beads (Sigma) as a carrier. During a 6-day period, bacterial growth was monitored daily with the Xenogen IVIS imaging system (Xenogen). For L. monocytogenes infection, mice were inoculated with logarithmic phase Listeria grown in brain-heart infusion broth at 37°C. Intravenous inoculation was performed via tail vein with 100 ml of bacteria in PBS. The inoculum was verified by counting colony-forming units after infection. Bioluminescence imaging was performed by using the IVIS Imaging System. Day 2 after infection, mice were anesthetized by isofluorane inhalation and shaved, and bioluminescence was recorded for 1 min at a pixel binning of 10. At day 3, light emission was measured for 10 s.Virus Culture, Infection, and Assay.
Mouse CMV (Smith strain) was prepared by in vivo propagation in 3-week-old BALB/c mice infected with 1 × 104 plaque-forming units by an i.p. route. Salivary gland homogenates were prepared at day 14 after infection, and viral titer was determined by plaque assay and by in vivo survival of BALB/c and C57BL/6 mice. Spleens were extracted on day 5 after mouse CMV infection, and three dilutions of their homogenates were used to determine viral titers by plaques.Constructs and Mutagenesis.
The N-terminal Flag-tag versions of murine TLR2, TLR4, and TLR6 expression vectors were made by removing the original signal peptides and replacing them with the signal peptide from preprotrypsin, followed by a 3XFLAG epitope tag in pCMV8 (Sigma). HA-tagged murine MyD88 and MyD88poc were cloned in pCMV. Human MD2 expression vector pEFBOS hMD2 was kindly provided by Sachiko Akashi Takamura (University of Tokyo, Tokyo). pNiFty-luc, an NF-kB-dependent E-selectin-luc reporter plasmid, were purchased from InvivoGen (San Diego). Point mutation of different TLR constructs were generated by the QuikChange site-directed mutagenesis kits with pfu-ultra as the polymerase (Stratagene) and the construct coding for the wild-type protein as the template.The primers used for mutagenesis are as follows.
For MyD88P200H:
5'-ACCGCGATGTCCTGCATGGCACCTGTGTCT
5'-AGACACAGGTGCCATGCAGGACATCGCGGT
For MyD88FW/AA:
5'-TGCACCAAATCTTGGGCCGCGACTCGCCTTGCCAAG
5'-CTTGGCAAGGCGAGTCGCGGCCCAAGATTTGGTGCA
For TLR2P681H:
5'-AAGCGGGACTTCGTTCATGGCAAATGGATCATT
5'-AATGATCCATTTGCCATGAACGAAGTCCCGCTT
For TLR2V660N:
5'-GTGGAGAACCTCATGAACCAGCAGCTGGAGAAC
5'-GTTCTCCAGCTGCTGGTTCATGAGGTTCTCCAC
For TLR2FW/AA:
5'-GGCCAGCAGGAAGTGGCTGCGGTAAATCTGAGAACTGCA
5'-TGCAGTTCTCAGATTTACCGCAGCCACTTCCTGCTGGCC
For TLR6P691H:
5'-CATGAGAGGAACTTTGTCCATGGCAAGAGCATTGTGGAG
5'-CTCCACAATGCTCTTGCCATGGACAAAGTTCCTCTCATG
For TLR6-L671N/L672N (named as mTLR6LL/NN):
5'-GCCTGGGTGAAGAACGAAAATAATCCCAACCTAGAGAAAGAT
5'-ATCTTTCTCTAGGTTGGGATTATTTTCGTTCTTCACCCAGGC
For hTLR4V693N:
5'-TAAGGAATGAGCTAAATAAGAATTTAGAAGAAG
5'-CTTCTTCTAAATTCTTATTTAGCTCATTCCTTA
For TLR9L891N:
5'-TGGGTGTATAACGAGAATCGGGTGCGGCTGGAGGAG
5'-CTCCTCCAGCCGCACCCGATTCTCGTTATACACCCA
To reverse the point mutation TLR4V693N back to the wild type, TLR4V693N was used as the template, and the following primers were used:
5'-TAAGGAATGAGCTAGTAAAGAATTTAGAAGAAG
5'-CTTCTTCTAAATTCTTTACTAGCTCATTCCTTA.
Each mutation was confirmed by sequencing.
1. Francis, K. P., Joh, D., Bellinger-Kawahara, C., Hawkinson, M. J., Purchio, T. F. & Contag, P. R. (2000) Infect. Immun. 68, 3594–3600.
2. Portnoy, D. A., Jacks, P. S. & Hinrichs, D. J. (1988) J. Exp. Med. 167, 1459–1471.